Intro to R
Module 3: Working with data
26 January 2024
Materials
Script
Click here to download the script! Save the script to the project directory you set up in the previous module.
Load your script in RStudio. To do this, open RStudio and click on the folder icon in the toolbar at the top to load your script.
Cheat sheets
Make sure to save these to your cheatsheets folder
Tidyverse
cheat sheet
readr
cheat sheet
tidyr and dplyr
cheat sheets Scroll down to data tidying with tidyr
& data transformation with dplyr
Working directory
Remember, the working directory is the first place R looks for any files you would like to read in (e.g., code, data). It is also the first place R will try to write any files you want to save.
Every R session has a working directory, whether you specify one or not. Find out what your working directory is.
# Working directories --------------------
# Find the directory you're working in
getwd() # note: the results from running this command on my machine will differ from yours! ## [1] "/Users/marissadyck/Documents/Teaching/ES482/ES482-R-labs.github.io"Since you should already be working in an RStudio Project, your working directory will be set as your project directory (directory that contains the .Rproj file for your project). This is convenient, because:
- That’s most likely where the data for that project live anyway
- If you’re collaborating with someone else on the project (e.g., in a shared Dropbox folder), you can both open the project and R will instantly know where to read and write data without either of you having to reset the working directory (even though the directory path is probably different on your two machines). This can save a lot of headaches!
If you didn’t open your Rstudio project before beginning today’s lab your working directory might show something different, if so close out of R and navigate to the project you created for this course and open Rstudio by double clicking the project.
Basic data exploration (base R)
Importing data into R
There are many ways data can be imported into R. Many types of files can be imported (e.g., text files, csv, shapefiles). And people are always inventing new ways to read and write data to/from R. But here are the basics.
- read.table or read_delim
- reads a data file in any major text format (comma-delimited, space
delimited etc.), you can specify which format (very general)
- reads a data file in any major text format (comma-delimited, space
delimited etc.), you can specify which format (very general)
- read.csv or read_csv
- fields are separated by a comma (this is still probably the most common way to read in data)
Before we can read data in, we need to put some data files in our working directory!
Download the following data files and store them in your ‘data’ folder in the R Crash Course working directory
Once you have saved these files to your working directory, open one or two of them up (e.g., in Excel or a text editor) to see what’s inside. You can do this directly in R using the files panel.
Now let’s read them into R!
# Import/Export data files into R with base R----------------------
# read.table to import text file (if no delim specified, it will try to guess!)
data.txt.df <- read.table("data/raw/data.txt") It is always useful to check what the data look like after they are read in. Can you recall one of the ways to view an object in R? View the data.txt.df object
# Look at data by printing it in the console
# You can also look at data by using the View() function
# Or you can click on the object (data.txt.df) in your environment to view it
data.txt.df## V1 V2 V3 V4
## 1 Country Import Export Product
## 2 Bolivia 46 0 N
## 3 Brazil 74 0 N
## 4 Chile 89 16 N
## 5 Colombia 77 16 A
## 6 CostaRica 84 21 A
## 7 Cuba 89 15 A
## 8 DominicanRep 68 14 A
## 9 Ecuador 70 6 A
## 10 ElSalvador 60 13 A
## 11 Guatemala 55 9 A
## 12 Haiti 35 3 A
## 13 Honduras 51 7 A
## 14 Jamaica 87 23 A
## 15 Mexico 83 4 N
## 16 Nicaragua 68 0 A
## 17 Panama 84 19 N
## 18 Paraguay 74 3 A
## 19 Peru 73 0 N
## 20 TrinidadTobago 84 15 A
## 21 Venezuela 91 7 AData will often be saved as a .csv file (or separated by commas) we can load this data using read.csv
# read.csv to import textfile with columns separated by commas
data.df <- read.csv("data/raw/data.csv")
#print data
data.df## Country Import Export Product
## 1 Bolivia 46 0 N
## 2 Brazil 74 0 N
## 3 Chile 89 16 N
## 4 Colombia 77 16 A
## 5 CostaRica 84 21 A
## 6 Cuba 89 15 A
## 7 DominicanRep 68 14 A
## 8 Ecuador 70 6 A
## 9 ElSalvador 60 13 A
## 10 Guatemala 55 9 A
## 11 Haiti 35 3 A
## 12 Honduras 51 7 A
## 13 Jamaica 87 23 A
## 14 Mexico 83 4 N
## 15 Nicaragua 68 0 A
## 16 Panama 84 19 N
## 17 Paraguay 74 3 A
## 18 Peru 73 0 N
## 19 TrinidadTobago 84 15 A
## 20 Venezuela 91 7 NRemoving objects from the environment
Since both of these files contain the same data just in different
formats let’s delete one of the from the environment so we don’t get
confused. We can use the rm() function for this.
# Remove redundant objects from memory
rm(data.txt.df)Built in data
R has many useful built-in data sets that come pre-loaded. You can explore these datasets with the following command:
# Using Rs built-in data----------------------
# Look at all built-in data files
data() Let’s read in one of these datasets!
# read built-in data on car road tests performed by Motor Trend
data(mtcars)
# inspect the first few lines
head(mtcars)
# learn more about this built-in data set
# ?mtcars Basic data checking
To learn more about the ‘internals’ of any data object in R, we can
use the str() (structure) function:
# Basic data checking ----------------------
# ?str: displays the internal structure of the data object
str(mtcars)## 'data.frame': 32 obs. of 11 variables:
## $ mpg : num 21 21 22.8 21.4 18.7 18.1 14.3 24.4 22.8 19.2 ...
## $ cyl : num 6 6 4 6 8 6 8 4 4 6 ...
## $ disp: num 160 160 108 258 360 ...
## $ hp : num 110 110 93 110 175 105 245 62 95 123 ...
## $ drat: num 3.9 3.9 3.85 3.08 3.15 2.76 3.21 3.69 3.92 3.92 ...
## $ wt : num 2.62 2.88 2.32 3.21 3.44 ...
## $ qsec: num 16.5 17 18.6 19.4 17 ...
## $ vs : num 0 0 1 1 0 1 0 1 1 1 ...
## $ am : num 1 1 1 0 0 0 0 0 0 0 ...
## $ gear: num 4 4 4 3 3 3 3 4 4 4 ...
## $ carb: num 4 4 1 1 2 1 4 2 2 4 ...str(data.df)## 'data.frame': 20 obs. of 4 variables:
## $ Country: chr "Bolivia" "Brazil" "Chile" "Colombia" ...
## $ Import : int 46 74 89 77 84 89 68 70 60 55 ...
## $ Export : int 0 0 16 16 21 15 14 6 13 9 ...
## $ Product: chr "N" "N" "N" "A" ...And we can use the summary() function to get a brief
summary of what’s in our data frame:
# get summary of the variables (columns) of a data object
summary(data.df)## Country Import Export Product
## Length:20 Min. :35.0 Min. : 0.00 Length:20
## Class :character 1st Qu.:66.0 1st Qu.: 3.00 Class :character
## Mode :character Median :74.0 Median : 8.00 Mode :character
## Mean :72.1 Mean : 9.55
## 3rd Qu.:84.0 3rd Qu.:15.25
## Max. :91.0 Max. :23.00If we want to look at data for a specific column in our dataset we
use the $ argument to specify which column
# get summary info for a specific variable
summary(data.df$Import)## Min. 1st Qu. Median Mean 3rd Qu. Max.
## 35.0 66.0 74.0 72.1 84.0 91.0Exporting data
Reading in data is one thing, but you will probably also want to write data to your hard drive as well. There are countless reasons for this- you might want to use an external program to plot your data, you might want to archive some simulation results, etc..
Again, there are many ways to write data to a file. Here are the basics using base R
# Exporting data (save to hard drive as data file)
# ?write.csv: writes a CSV file to the working directory
# to save an entire data frame to your hard drive you would use the format below
# write.csv(data name in R, file = 'data name to save as.csv')
# however if we did this for data.df we would just be making a copy, instead we can export a subset of the data we just ran in
write.csv(data.df[, c("Country","Product")], # this tells R to save all rows for the 'Country' and 'Product' columns
file = "data/processed/data_export.csv") Working with data (base R)
Now let’s start seeing what we can do with data in R. Even without doing any statistical analyses, R is very a powerful environment for doing data transformations and performing mathematical operations.
Boolean operations
Boolean operations refer to TRUE/FALSE tests. That is, we can ask a question about the data to a Boolean operator and the operator will return a TRUE or a FALSE (logical) result.
First, let’s meet the boolean operators.
NOTE: don’t get confused between the equals sign (=),
which is an assignment operator (same as <-),
and the double equals sign (==), which is a Boolean
operator:
# Working with data in R ------------------------
# Assigning objects ----------------------
# <- assignment operator
# = alternative assignment operator *avoid using this to assign objects to the environment
a <- 3 # assign the value "3" to the object named "a"
a = 3 # assign the value "3" to the object named "a" (alternative)
a == 3 # answer the question: "does the object "a" equal "3"? ## [1] TRUE # Boolean operations ----------------------
# Basic operators
# < less than
# > greater than
# <= less than or equal to
# >= greater than or equal to
# == equal to
# != not equal to
# %in% matches one of a specified group of possibilities
# Combining multiple conditions
# & must meet both conditions (AND operator)
# | must meet one of two conditions (OR operator)
Y <- 4
Z <- 6
Y == Z #I am asking if Y is equal to Z, and it will return FALSE
Y < Z
!(Y < Z) # the exclamation point reverses any boolean object (read "NOT")
# Wrong!
data.df[, 2] = 74 # sets entire second column equal to 74! OOPS WE GOOFED UP!!!
data.df <- read.csv("data/data.csv") ## correct our mistake in the previous line (revert to the original data)!
# Right
data.df[, 2] == 74 # tests each element of column to see whether it is equal to 74
data.df[, 2] < 74 | data.df[, 2] == 91 # use the logical OR operatorThis image from Wickham and Grolemund’s excellent R for Data Science book can help conceptualize the combination operators:
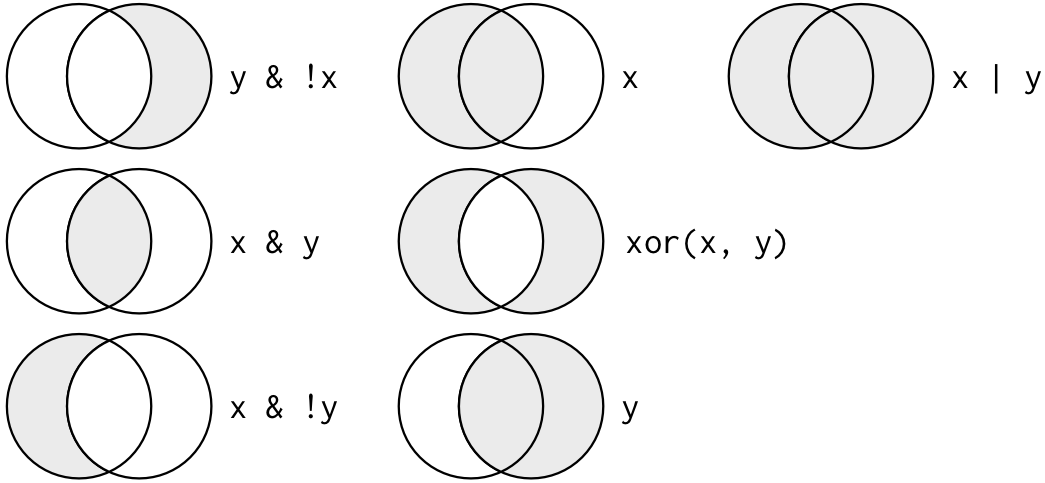
Data subsetting using boolean logic
You probably won’t us boolean operations that often to return TRUE/FALSE results but they are great for subsetting, filtering, and reformatting data.
Let’s use them to subset data - select only those rows/observations that meet a certain condition (where [some condition] is TRUE).
# Sub-setting data base R----------------------
# select those rows of data for which second column is less than 74
data.df[data.df[, "Import"] < 74, ] ## Country Import Export Product
## 1 Bolivia 46 0 N
## 7 DominicanRep 68 14 A
## 8 Ecuador 70 6 A
## 9 ElSalvador 60 13 A
## 10 Guatemala 55 9 A
## 11 Haiti 35 3 A
## 12 Honduras 51 7 A
## 15 Nicaragua 68 0 A
## 18 Peru 73 0 N# select rows of data for which second column is greater than 25 AND less than 75
data.df[data.df[, 'Import'] > 25 &
data.df[, 'Import'] <75, ]## Country Import Export Product
## 1 Bolivia 46 0 N
## 2 Brazil 74 0 N
## 7 DominicanRep 68 14 A
## 8 Ecuador 70 6 A
## 9 ElSalvador 60 13 A
## 10 Guatemala 55 9 A
## 11 Haiti 35 3 A
## 12 Honduras 51 7 A
## 15 Nicaragua 68 0 A
## 17 Paraguay 74 3 A
## 18 Peru 73 0 NSummary statistics
We can also use base R functions to get summary
statistics for our data. Let’s use the functions
mean() and sd() to calculate the mean and
standard deviation for one of the columns in our data frame.
# Summary statistics base R ----------------------
mean(data.df$Import)## [1] 72.1sd(data.df$Import)## [1] 15.73765However, if we wanted to calculate the mean and sd for many columns in our dataset this would take a lot of lines of repetitive code using base R. We will go over a much more efficient way using the tidyverse package.
Packages
Packages are an incredibly useful to for R users. These are extensions of the R programming language which contain a vast array of functions, code, sample data etc. A few rules for using packages;
Have to be installed (once but you may need to re-install the latest version as the packages are updated)
Loaded into your library (every time you start a new R session). The library is simply the directory where the packages are stored.
Installing packages
Let’s install some packages that we will use for this module. There are a few ways to do this
Use the packages panel>>install>>type the package you want
Use the
install.packages()function.
# Packages----------------------
# Install packages
# install.packages("tidyverse")*Notice that the package name is enclosed with quotations (” “). If you do not use the quotations the package will not be installed.
Loading packages to library
Each time you open a new R session you need to load the
packages you want to use into your
library. To do this use the library()
function
# Load packages to library
library(tidyverse)Built-in datasets
Many packages come with built-in data sets as well. For, instance, ggplot2 comes with the “diamonds” data:
# Package datasets ----------------------
ggplot2::diamonds # note the use of the package name followed by two colons- this is a way to make sure you are using a function (or data set or other object) from a particular package... (sometimes several packages have functions with the same name...)## # A tibble: 53,940 × 10
## carat cut color clarity depth table price x y z
## <dbl> <ord> <ord> <ord> <dbl> <dbl> <int> <dbl> <dbl> <dbl>
## 1 0.23 Ideal E SI2 61.5 55 326 3.95 3.98 2.43
## 2 0.21 Premium E SI1 59.8 61 326 3.89 3.84 2.31
## 3 0.23 Good E VS1 56.9 65 327 4.05 4.07 2.31
## 4 0.29 Premium I VS2 62.4 58 334 4.2 4.23 2.63
## 5 0.31 Good J SI2 63.3 58 335 4.34 4.35 2.75
## 6 0.24 Very Good J VVS2 62.8 57 336 3.94 3.96 2.48
## 7 0.24 Very Good I VVS1 62.3 57 336 3.95 3.98 2.47
## 8 0.26 Very Good H SI1 61.9 55 337 4.07 4.11 2.53
## 9 0.22 Fair E VS2 65.1 61 337 3.87 3.78 2.49
## 10 0.23 Very Good H VS1 59.4 61 338 4 4.05 2.39
## # ℹ 53,930 more rowsTidyverse
The tidyverse is a collection of packages introduced by Hadley and Wickham that all work together using a similar design, syntax, and data structure (tidy data) for data science. It will be useful to have your tidyverse cheat sheet, reader cheat sheet, and tidyr & dplyr cheat sheets during the rest of this module.
What is tidy data
All of the tidyverse set of packages are designed to work with Tidy formatted data.
This means:
- Each variable must have its own column.
- Each observation must have its own row.
- Each value must have its own cell.
This is what it looks like:
 We will start by working with data
that is already ‘tidy’ but later we will cover several
functions to help you get your data into this
format.
We will start by working with data
that is already ‘tidy’ but later we will cover several
functions to help you get your data into this
format.
Base R vs tidyverse
We didn’t spend a lot of time going over data exploration, manipulation, or visualization in base R because the tidyverse provides a much more efficient and intuitive way to do these things. I will provide a few examples as we continue to compare base R to the tidyverse to illustrate this point.
Importing tidy data (tibbles)
We already covered a few ways to read in data with base
R but since we will primarily be using the
tidyverse for this course let’s cover the tidy
functions to read in data. The
function to read in .txt files as tidy data is
read_delim() in the readr
package.
# The wonders of tidyverse ----------------------
# Read in tidy data ----------------------
tidy_data.txt <- read_delim('data/raw/data.txt')
# print data
tidy_data.txt## # A tibble: 20 × 4
## Country Import Export Product
## <chr> <dbl> <dbl> <chr>
## 1 Bolivia 46 0 N
## 2 Brazil 74 0 N
## 3 Chile 89 16 N
## 4 Colombia 77 16 A
## 5 CostaRica 84 21 A
## 6 Cuba 89 15 A
## 7 DominicanRep 68 14 A
## 8 Ecuador 70 6 A
## 9 ElSalvador 60 13 A
## 10 Guatemala 55 9 A
## 11 Haiti 35 3 A
## 12 Honduras 51 7 A
## 13 Jamaica 87 23 A
## 14 Mexico 83 4 N
## 15 Nicaragua 68 0 A
## 16 Panama 84 19 N
## 17 Paraguay 74 3 A
## 18 Peru 73 0 N
## 19 TrinidadTobago 84 15 A
## 20 Venezuela 91 7 ANotice the tidy way did a slightly better job reading in this data (e.g., we don’t have those weird variable labels at the top of our columns)
You’ll also notice the object type has change to a tibble
# check the object type of data
class(tidy_data.txt)## [1] "spec_tbl_df" "tbl_df" "tbl" "data.frame"The function to read in .csv files as tidy data is
read_csv() in the readr
package
tidy_data.csv <- read_csv('data/raw/data.csv')
# print the first few rows of data
head(tidy_data.csv)## # A tibble: 6 × 4
## Country Import Export Product
## <chr> <dbl> <dbl> <chr>
## 1 Bolivia 46 0 N
## 2 Brazil 74 0 N
## 3 Chile 89 16 N
## 4 Colombia 77 16 A
## 5 CostaRica 84 21 A
## 6 Cuba 89 15 AWe have now imported multiple versions of the same data with similar names. Let’s remove all but one from our environment to avoid confusion later on. Recall the command to remove objects from your environment? using the console remove ‘tidy_data.txt’ and ‘data.df’ from your environment.
# in the console type
# rm(tidy_data.txt)
# rm(data.df)Exporting tidy data
The function to save tidy data as .csv files is
write_csv() in the readr
package
# Export tidy data ----------------------
# ?write_csv
# write_csv(data, 'data_name_to_save.csv')Check your data import (readr) cheat sheet for more functions to import and export tidy data*
The pipe operator (%>%)
The tidyverse set of packages takes
advantage of the pipe operator %>%,
which provides a clean and intuitive way to structure code and perform
sequential operations in R.
 %>%
%>%
Key advantages include:
code is more readable and intuitive – reads left to right, rather than inside out as is the case for nested function
perform multiple operations without creating a bunch of intermediate (temporary) datasets
This operator comes from the magrittr package, which is included in the installation of all of the tidyverse packages. The shortcut for the pipe operator is ctrl + shift + m (‘m’ is for Magritte) for Windows and command + shift + m for Mac. When reading code out loud, use ‘then’ for the pipe. For example the command here:
x %>%
log() %>%
round(digits = 2)
can be interpreted as follows:
Take “x”, THEN take its natural logarithm, THEN round the resulting value to 2 decimal places
The structure is simple. Start with the object you want to manipulate, and apply actions (e.g., functions) to that object in the order in which you want to apply them.
Here is a quick example of dplyr in action using the dataset we read in earlier.
# The pipe operator ----------------------
# start by assigning a value to x
x <- 3
# call the variable x followed by %>% ('then')
x %>%
# calculate the log of x, 'then'
log() %>%
# round the log of x to 2 digits
round(digits = 2) ## [1] 1.1For the above code to be applied in base R you would have to nest your functions which can get very confusing to read or assign many objects to your environment that you only need temporarily. For example,
x <- 3
# nested code
round(log(x), digits=2) ## [1] 1.1# assigning multiple objects
log_x <- log(x)
round_log_x <- round(x, digits = 2)The nested code may seem simpler at first but you can imagine as operation get more complex this can become quite difficult to interpret.
Data exploration (tidyverse)
Let’s switch to a different data file. Download the Turtle Data and save it to the data folder in your project directory.
Let’s use this data set to review tsome data processing operations we have learned- and learn a few new things along the way:
First we need to read the data in to R. Using the reader package, read the turtle_data.txt in to R and assign it to the environemnt as turtles.df
# Data exploration with tidyverse ----------------------
# read turtle_data.txt in to R using readr
turtles.df <- read_delim('data/raw/turtle_data.txt',
delim = '\t') # specify tab-delimited fileCheck data
It is always good coding practice to view your data and check the internal structure before proceeding with any processing, analyses, or visualization. View the turtles, check the internal structure and then look at a summary of the columns. Below are the outputs you should see in the console.
## spc_tbl_ [21 × 5] (S3: spec_tbl_df/tbl_df/tbl/data.frame)
## $ Tag_number : num [1:21] 10 11 2 15 16 3 4 5 12 13 ...
## $ Sex : chr [1:21] "male" "female" NA NA ...
## $ Carapace_length: num [1:21] 41 46.4 24.3 28.7 32 42.8 40 45 44 28 ...
## $ Head_width : num [1:21] 7.15 8.18 4.42 4.89 5.37 7.32 6.6 8.05 7.55 4.85 ...
## $ Weight : num [1:21] 7.6 11 1.65 2.18 3 8.6 6.5 10.9 8.9 1.97 ...
## - attr(*, "spec")=
## .. cols(
## .. Tag_number = col_double(),
## .. Sex = col_character(),
## .. Carapace_length = col_double(),
## .. Head_width = col_double(),
## .. Weight = col_double()
## .. )
## - attr(*, "problems")=<externalptr>## Tag_number Sex Carapace_length Head_width
## Min. : 1.00 Length:21 Min. :24.30 Min. :4.42
## 1st Qu.: 6.00 Class :character 1st Qu.:32.00 1st Qu.:5.37
## Median : 11.00 Mode :character Median :41.00 Median :6.60
## Mean : 19.19 Mean :38.71 Mean :6.58
## 3rd Qu.: 16.00 3rd Qu.:44.00 3rd Qu.:7.35
## Max. :105.00 Max. :48.10 Max. :8.67
## Weight
## Min. : 1.650
## 1st Qu.: 3.000
## Median : 7.200
## Mean : 6.737
## 3rd Qu.: 9.000
## Max. :13.500Renaming columns
Notice the column names are capitalized in this data set. Data are often entered this way, but this can lead to mistakes in your code if you forget to capitalize a variable. I prefer to keep everything lowercase when working in R to avoid this and extra keystrokes. There’s an easy way to do this using the dplyr package.
# Renaming columns ----------------------
# check names of turtles data before changing
names(turtles.df)## [1] "Tag_number" "Sex" "Carapace_length" "Head_width"
## [5] "Weight"turtles.df %>%
# set column names to lowercase
set_names(
names(.) %>% # the period here is a placeholder for the data it tells R to use the element before the last pipe (e.g., turtles.df)
tolower()) ## # A tibble: 21 × 5
## tag_number sex carapace_length head_width weight
## <dbl> <chr> <dbl> <dbl> <dbl>
## 1 10 male 41 7.15 7.6
## 2 11 female 46.4 8.18 11
## 3 2 <NA> 24.3 4.42 1.65
## 4 15 <NA> 28.7 4.89 2.18
## 5 16 <NA> 32 5.37 3
## 6 3 female 42.8 7.32 8.6
## 7 4 male 40 6.6 6.5
## 8 5 female 45 8.05 10.9
## 9 12 female 44 7.55 8.9
## 10 13 <NA> 28 4.85 1.97
## # ℹ 11 more rows# check the names of turtles data
names(turtles.df)## [1] "Tag_number" "Sex" "Carapace_length" "Head_width"
## [5] "Weight"Alternatively a more efficient way to do this would be to change the names in the same code chunk that we read in the data. Let’s check this, remove the turtles data from your environment.
rm(turtles.df)Now let’s read in the data and change the column names to lowercase in the same code chunk.
# read turtle_data.txt in to R using readr
turtles.df <- read_delim('data/raw/turtle_data.txt',
delim = '\t') %>%
# set column names to lowercase
set_names(
names(.) %>%
tolower())
summary(turtles.df)## tag_number sex carapace_length head_width
## Min. : 1.00 Length:21 Min. :24.30 Min. :4.42
## 1st Qu.: 6.00 Class :character 1st Qu.:32.00 1st Qu.:5.37
## Median : 11.00 Mode :character Median :41.00 Median :6.60
## Mean : 19.19 Mean :38.71 Mean :6.58
## 3rd Qu.: 16.00 3rd Qu.:44.00 3rd Qu.:7.35
## Max. :105.00 Max. :48.10 Max. :8.67
## weight
## Min. : 1.650
## 1st Qu.: 3.000
## Median : 7.200
## Mean : 6.737
## 3rd Qu.: 9.000
## Max. :13.500Sometimes we have column names that are not informative or don’t make
sense to us, and rather than just changing them to lowercase we may want
to rename some of them. Let’s go through how to rename columns using the
names() function. Let’s make some shorter
names for the turtles dataset.
# extract the column names for turtle data
names(turtles.df)## [1] "tag_number" "sex" "carapace_length" "head_width"
## [5] "weight"# to change the column names, use the "names()" function
names(turtles.df) <- c("tag", "sex","c_length", "h_width", "weight")
# let's check
head(turtles.df)## # A tibble: 6 × 5
## tag sex c_length h_width weight
## <dbl> <chr> <dbl> <dbl> <dbl>
## 1 10 male 41 7.15 7.6
## 2 11 female 46.4 8.18 11
## 3 2 <NA> 24.3 4.42 1.65
## 4 15 <NA> 28.7 4.89 2.18
## 5 16 <NA> 32 5.37 3
## 6 3 female 42.8 7.32 8.6We can also do this more efficiently in the dplyr pipe when
we read in the data using the rename()
function.
# read turtle_data.txt in to R using readr
turtles.df <- read_delim('data/raw/turtle_data.txt',
delim = '\t') %>%
# set column names to lowercase
set_names(
names(.) %>%
tolower()) %>%
# rename columns to shorter names
rename(tag = tag_number,
c_length = carapace_length,
h_width = head_width)
summary(turtles.df)## tag sex c_length h_width
## Min. : 1.00 Length:21 Min. :24.30 Min. :4.42
## 1st Qu.: 6.00 Class :character 1st Qu.:32.00 1st Qu.:5.37
## Median : 11.00 Mode :character Median :41.00 Median :6.60
## Mean : 19.19 Mean :38.71 Mean :6.58
## 3rd Qu.: 16.00 3rd Qu.:44.00 3rd Qu.:7.35
## Max. :105.00 Max. :48.10 Max. :8.67
## weight
## Min. : 1.650
## 1st Qu.: 3.000
## Median : 7.200
## Mean : 6.737
## 3rd Qu.: 9.000
## Max. :13.500Summary statistics
Remember earlier we calculated the mean and standard deviation for a column in our dataset? Calculate the mean and sd for carapace length, head width, and weight in the turtles data.
# Summary stats the tidyverse way ----------------------
# calculate the mean and sd for three columns in the turtles data using base R
# carapace length
mean(turtles.df$c_length)## [1] 38.70952sd (turtles.df$c_length)## [1] 7.223427# head width
mean(turtles.df$h_width)## [1] 6.58sd(turtles.df$h_width)## [1] 1.272108# weight
mean(turtles.df$weight)## [1] 6.737143sd(turtles.df$weight)## [1] 3.666874We had to type a lot to do these simple calculations across multiple
columns, and there is a lot of repetition in our code which we should
try to avoid. And what if we wanted a simple table with the meand and sd
for all three columns so we could easily compare. This would involved a
lot more coding. Luckily, the tidyverse dplyr
package allows for a much more efficient way to do this
using the summarise() function.
# calculate the mean and sd for three columns in the turtles data using dplyr
turtles.df %>%
# use summarise function to get mean and sd for multiple columns
summarise(cl_mean = mean(c_length),
cl_sd = sd(c_length),
hw_mean = mean(h_width),
hw_sd = sd(h_width),
w_mean = mean(weight),
w_sd = sd(weight)
)## # A tibble: 1 × 6
## cl_mean cl_sd hw_mean hw_sd w_mean w_sd
## <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 38.7 7.22 6.58 1.27 6.74 3.67Using dplyr we only have to reference the data once which saves us
typing and our output is a nice tibble data frame that is easy to read
or save and use later for plotting etc. However, there’s still a more
efficient way to do this in dplyr let’s try the
summarise_all() function.
turtles.df %>%
summarise_all(.funs = list(mean, sd))## # A tibble: 1 × 10
## tag_fn1 sex_fn1 c_length_fn1 h_width_fn1 weight_fn1 tag_fn2 sex_fn2
## <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 19.2 NA 38.7 6.58 6.74 28.9 NA
## # ℹ 3 more variables: c_length_fn2 <dbl>, h_width_fn2 <dbl>, weight_fn2 <dbl>This worked but since a few of our variables are not
numeric R cannot calculate a mean and sd for them and
we get NAs. Let’s try summarise_if() instead and specify
only to run the functions if the variables are
numeric.
turtles.df %>%
summarise_if(is.numeric,
.funs = list(mean, sd))## # A tibble: 1 × 8
## tag_fn1 c_length_fn1 h_width_fn1 weight_fn1 tag_fn2 c_length_fn2 h_width_fn2
## <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 19.2 38.7 6.58 6.74 28.9 7.22 1.27
## # ℹ 1 more variable: weight_fn2 <dbl>This worked better but the column names for our new
tibble aren’t very informative if we had a lot of
functions to run it would be hard to remember which is which. Let’s try
adding names. Let’s practice using the help window to
see if we can figure out how to name our functions.
Use the console to access
the R documentation for
summarise_if().
# ?summarise_if()Let’s work through the R documentation together. It
looks like there is a ‘naming’ section for this
function that explains exactly how the columns are
named based on the arguments/input you provide in the
function, but I don’t see how to specify the column
names… Let’s see if some of the examples help us. It looks like the very
last one provides names for each function in the list()
argument. Let’s copy this code to the console and run
it to check if the output is what we want.
by_species <- iris %>%
group_by(Species)
by_species %>%
summarise(across(everything(), list(min = min, max = max)))## # A tibble: 3 × 9
## Species Sepal.Length_min Sepal.Length_max Sepal.Width_min Sepal.Width_max
## <fct> <dbl> <dbl> <dbl> <dbl>
## 1 setosa 4.3 5.8 2.3 4.4
## 2 versicolor 4.9 7 2 3.4
## 3 virginica 4.9 7.9 2.2 3.8
## # ℹ 4 more variables: Petal.Length_min <dbl>, Petal.Length_max <dbl>,
## # Petal.Width_min <dbl>, Petal.Width_max <dbl>Looks good! Let’s try this with our data. Using our code from earlier and the example above, get the mean and sd for all numeric variables in the turtle data and specify the names of the functions ‘mean’ and ‘sd’.
turtles.df %>%
summarise_if(is.numeric,
.funs = list(mean = mean,
sd = sd))## # A tibble: 1 × 8
## tag_mean c_length_mean h_width_mean weight_mean tag_sd c_length_sd h_width_sd
## <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 19.2 38.7 6.58 6.74 28.9 7.22 1.27
## # ℹ 1 more variable: weight_sd <dbl>This is much better! And you just practiced using the help window and R documentation to learn about a new function
Change variable type
In the last example we specified to only calculate summary statistics
for variables that were numeric. But what if R doesn’t
properly read the variables types and we want to change them? This
happens quite frequent, let’s look at how to do this in dplyr
using the mutate() function. This
function creates new columns using existing variables
and can also modify existing columns by settting the new column name the
same as a previous column name, as in this example below.
# Change variable type ----------------------
# Change sex to factor
turtles.df %>%
mutate(sex = as.factor(sex)) # we use the same name for our 'new' column so that it modifies the old column instead of creating a new one## # A tibble: 21 × 5
## tag sex c_length h_width weight
## <dbl> <fct> <dbl> <dbl> <dbl>
## 1 10 male 41 7.15 7.6
## 2 11 female 46.4 8.18 11
## 3 2 <NA> 24.3 4.42 1.65
## 4 15 <NA> 28.7 4.89 2.18
## 5 16 <NA> 32 5.37 3
## 6 3 female 42.8 7.32 8.6
## 7 4 male 40 6.6 6.5
## 8 5 female 45 8.05 10.9
## 9 12 female 44 7.55 8.9
## 10 13 <NA> 28 4.85 1.97
## # ℹ 11 more rows# check structure
str(turtles.df) # sex is still a character, why?## spc_tbl_ [21 × 5] (S3: spec_tbl_df/tbl_df/tbl/data.frame)
## $ tag : num [1:21] 10 11 2 15 16 3 4 5 12 13 ...
## $ sex : chr [1:21] "male" "female" NA NA ...
## $ c_length: num [1:21] 41 46.4 24.3 28.7 32 42.8 40 45 44 28 ...
## $ h_width : num [1:21] 7.15 8.18 4.42 4.89 5.37 7.32 6.6 8.05 7.55 4.85 ...
## $ weight : num [1:21] 7.6 11 1.65 2.18 3 8.6 6.5 10.9 8.9 1.97 ...
## - attr(*, "spec")=
## .. cols(
## .. Tag_number = col_double(),
## .. Sex = col_character(),
## .. Carapace_length = col_double(),
## .. Head_width = col_double(),
## .. Weight = col_double()
## .. )
## - attr(*, "problems")=<externalptr>You’ll notice it works within you pipe but when we re-check the structure of our data it hasn’t changed. This is because we save the changes, to do this we need to use the assignment operator to save the altered turtles.df data to the environment. Using the same name for the turtles data and the assignment operator and code above assign the altered data to the environment as turtles.df. Check the structure of the data when you are done to ensure it worked.
# Change sex to factor as save changes to data
turtles.df <- turtles.df %>%
mutate(sex = as.factor(sex))
# check structure
str(turtles.df) ## tibble [21 × 5] (S3: tbl_df/tbl/data.frame)
## $ tag : num [1:21] 10 11 2 15 16 3 4 5 12 13 ...
## $ sex : Factor w/ 3 levels "fem","female",..: 3 2 NA NA NA 2 3 2 2 NA ...
## $ c_length: num [1:21] 41 46.4 24.3 28.7 32 42.8 40 45 44 28 ...
## $ h_width : num [1:21] 7.15 8.18 4.42 4.89 5.37 7.32 6.6 8.05 7.55 4.85 ...
## $ weight : num [1:21] 7.6 11 1.65 2.18 3 8.6 6.5 10.9 8.9 1.97 ...This worked! But, you guessed it. There is a more efficient way to do this right when we read in the data. Using the code below, add the mutate function to the dplyr pipe to mutate sex to a factor.
# read turtle_data.txt in to R using readr
turtles.df <- read_delim('data/turtle_data.txt',
delim = '\t') %>%
# set column names to lowercase
set_names(
names(.) %>%
tolower()) %>%
# rename columns to shorter names
rename(tag = tag_number,
c_length = carapace_length,
h_width = head_width)# read turtle_data.txt in to R using readr
turtles.df <- read_delim('data/raw/turtle_data.txt',
delim = '\t') %>%
# set column names to lowercase
set_names(
names(.) %>%
tolower()) %>%
# rename columns to shorter names
rename(tag = tag_number,
c_length = carapace_length,
h_width = head_width) %>%
# change sex to factor
mutate(sex = as.factor(sex))Similar to the summarise() function we
can also perform mutations on all the columns
(mutate_all()) or multiple columns that fit a criteria
(mutat_if()). Without assigning
the object to the environment to mutate all the
numeric columns to characters
in the turtles data. HINT: Look at the R Documentation
examples.
# read turtle_data.txt in to R using readr
turtles.df %>%
mutate_if(is.numeric, as.character)## # A tibble: 21 × 5
## tag sex c_length h_width weight
## <chr> <fct> <chr> <chr> <chr>
## 1 10 male 41 7.15 7.6
## 2 11 female 46.4 8.18 11
## 3 2 <NA> 24.3 4.42 1.65
## 4 15 <NA> 28.7 4.89 2.18
## 5 16 <NA> 32 5.37 3
## 6 3 female 42.8 7.32 8.6
## 7 4 male 40 6.6 6.5
## 8 5 female 45 8.05 10.9
## 9 12 female 44 7.55 8.9
## 10 13 <NA> 28 4.85 1.97
## # ℹ 11 more rowsSubsetting data with tidyr
Just like in base R we can use functions from the tidyverse packages to subset our data.
Filter
For example, if we are only interested in the male data we can use
the filter() function to do this with
Boolean operations. Filter applies criteria to the rows and returns only
the rows that match the criteria.
# Subsetting data with tidyr ----------------------
# Filter ----------------------
# subset turtles data
turtles.df %>%
# return data for males only
filter(sex == 'male')## # A tibble: 7 × 5
## tag sex c_length h_width weight
## <dbl> <fct> <dbl> <dbl> <dbl>
## 1 10 male 41 7.15 7.6
## 2 4 male 40 6.6 6.5
## 3 9 male 35 5.74 3.9
## 4 19 male 42.3 6.77 7.8
## 5 105 male 44 7.1 9
## 6 14 male 43 6.6 7.2
## 7 104 male 44 7.35 9turtles.df %>%
# return data for males only that are larger than 5 (grams?)
filter(sex == 'male' &
weight > 5)## # A tibble: 6 × 5
## tag sex c_length h_width weight
## <dbl> <fct> <dbl> <dbl> <dbl>
## 1 10 male 41 7.15 7.6
## 2 4 male 40 6.6 6.5
## 3 19 male 42.3 6.77 7.8
## 4 105 male 44 7.1 9
## 5 14 male 43 6.6 7.2
## 6 104 male 44 7.35 9Select
If we only want to work with specific columns we can also subset the
data using the select() function. We don’t
need Boolean operations here, instead we can just refer to the column
names.
# Select ----------------------
turtles.df %>%
# return data for tag and weight only
select(tag, weight)## # A tibble: 21 × 2
## tag weight
## <dbl> <dbl>
## 1 10 7.6
## 2 11 11
## 3 2 1.65
## 4 15 2.18
## 5 16 3
## 6 3 8.6
## 7 4 6.5
## 8 5 10.9
## 9 12 8.9
## 10 13 1.97
## # ℹ 11 more rowsWe can also combine these functions to subset the columns and rows in one code chunk.
turtles.df %>%
# return data for males only with weight greater than 5
filter(sex == 'male' &
weight > 5) %>%
# return data for tag and weight only
select(tag, weight)## # A tibble: 6 × 2
## tag weight
## <dbl> <dbl>
## 1 10 7.6
## 2 4 6.5
## 3 19 7.8
## 4 105 9
## 5 14 7.2
## 6 104 9And we can assign the altered data to the environment to save for later or to export to our hard drive if we want.
turtles_males_lg <- turtles.df %>%
# return data for males only with weight greater than 5
filter(sex == 'male' &
weight > 5) %>%
# return data for tag and weight only
select(tag, weight)Group by
group_by() is a useful function to get
summary statistics for various groups such as different treatments,
sexes, etc. You simply supply the column name in the
group_by() function and anything after
will be applied to each group separately. For example:
# Group by ----------------------
turtles.df %>%
# group by sex
group_by(sex) %>%
summarise(mean = mean(weight))## # A tibble: 4 × 2
## sex mean
## <fct> <dbl>
## 1 fem 6.2
## 2 female 10.0
## 3 male 7.29
## 4 <NA> 2.35 # END MAIN SCRIPT ----------------------You might notice a problem with the way the data are entered here. We will fix that in the next module.
Practice Problems
1 Export data
Save the altered turtles data as a comma separated file to the data/processed folder in your working directory using the ‘readr’ package and name it ‘turtles_tidy’
2 Import and rename data
Download the brown bear damage data (bear_2008_2016.csv) and save it in the data folder.
- Import the brown bear dataset from the data/raw folder
using the appropriate function from the ‘readr’ package and save it as
bear_data.
- In the same code chunk set the column names to lowercase and
- change dist_to_town to m_to_town and dist_to_forest to m_to_forest
(m for meters)
- Finally, use one of the functions we’ve covered to view/print your data to check that it worked.
## # A tibble: 6 × 25
## damage year month targetspp point_x point_y bear_abund landcover_code
## <dbl> <dbl> <dbl> <chr> <dbl> <dbl> <dbl> <dbl>
## 1 0 2008 0 <NA> 542583. 532675. 38 311
## 2 0 2008 0 <NA> 540963. 528113. 40 311
## 3 0 2008 0 <NA> 542435. 510385. 38 211
## 4 0 2008 0 <NA> 541244. 507297. 38 231
## 5 0 2008 0 <NA> 542791. 497755. 32 211
## 6 0 2008 0 <NA> 544639. 610454. 32 313
## # ℹ 17 more variables: altitude <dbl>, human_population <dbl>,
## # m_to_forest <dbl>, m_to_town <dbl>, livestock_killed <dbl>,
## # numberdamageperplot <dbl>, shannondivindex <dbl>, prop_arable <dbl>,
## # prop_orchards <dbl>, prop_pasture <dbl>, prop_ag_mosaic <dbl>,
## # prop_seminatural <dbl>, prop_deciduous <dbl>, prop_coniferous <dbl>,
## # prop_mixedforest <dbl>, prop_grassland <dbl>, prop_for_regen <dbl>3 Change variable type
- Check the internal structure of the bear dataset
- Change ‘targetspp’ from character to
factor using dplyr make sure to overwrite the
previous bear data so the changes are saved
- Use the ‘levels()’ function to check if this worked. HINT: Use the help file if you aren’t familiar with ‘levels()’
## spc_tbl_ [3,024 × 25] (S3: spec_tbl_df/tbl_df/tbl/data.frame)
## $ damage : num [1:3024] 0 0 0 0 0 0 0 0 0 0 ...
## $ year : num [1:3024] 2008 2008 2008 2008 2008 ...
## $ month : num [1:3024] 0 0 0 0 0 0 0 0 0 0 ...
## $ targetspp : chr [1:3024] NA NA NA NA ...
## $ point_x : num [1:3024] 542583 540963 542435 541244 542791 ...
## $ point_y : num [1:3024] 532675 528113 510385 507297 497755 ...
## $ bear_abund : num [1:3024] 38 40 38 38 32 32 37 45 45 36 ...
## $ landcover_code : num [1:3024] 311 311 211 231 211 313 313 312 324 312 ...
## $ altitude : num [1:3024] 800 666 504 553 470 ...
## $ human_population : num [1:3024] 2 0 2 0 0 0 0 0 0 0 ...
## $ m_to_forest : num [1:3024] 0 0 1829 398 942 ...
## $ m_to_town : num [1:3024] 2398 2441 571 1598 1068 ...
## $ livestock_killed : num [1:3024] 0 0 0 0 0 0 0 0 0 0 ...
## $ numberdamageperplot: num [1:3024] 0 0 0 0 0 0 0 0 0 0 ...
## $ shannondivindex : num [1:3024] 0.963 1.119 1.056 1.506 1.067 ...
## $ prop_arable : num [1:3024] 0 0 65.6 27.3 53.2 ...
## $ prop_orchards : num [1:3024] 0 0 0 0 0 0 0 0 0 0 ...
## $ prop_pasture : num [1:3024] 0 25.3 11.13 20 4.27 ...
## $ prop_ag_mosaic : num [1:3024] 0 0 8.55 0 0 ...
## $ prop_seminatural : num [1:3024] 16.3 30.6 0 19.8 0 ...
## $ prop_deciduous : num [1:3024] 57.796 43.095 0 0.769 32.454 ...
## $ prop_coniferous : num [1:3024] 0 0 0 0 0 ...
## $ prop_mixedforest : num [1:3024] 0 0 0 0 0 ...
## $ prop_grassland : num [1:3024] 0 1 0 0 0 ...
## $ prop_for_regen : num [1:3024] 25.9 0 13.5 0 0 ...
## - attr(*, "spec")=
## .. cols(
## .. Damage = col_double(),
## .. Year = col_double(),
## .. Month = col_double(),
## .. Targetspp = col_character(),
## .. POINT_X = col_double(),
## .. POINT_Y = col_double(),
## .. Bear_abund = col_double(),
## .. Landcover_code = col_double(),
## .. Altitude = col_double(),
## .. Human_population = col_double(),
## .. Dist_to_forest = col_double(),
## .. Dist_to_town = col_double(),
## .. Livestock_killed = col_double(),
## .. Numberdamageperplot = col_double(),
## .. ShannonDivIndex = col_double(),
## .. prop_arable = col_double(),
## .. prop_orchards = col_double(),
## .. prop_pasture = col_double(),
## .. prop_ag_mosaic = col_double(),
## .. prop_seminatural = col_double(),
## .. prop_deciduous = col_double(),
## .. prop_coniferous = col_double(),
## .. prop_mixedforest = col_double(),
## .. prop_grassland = col_double(),
## .. prop_for_regen = col_double()
## .. )
## - attr(*, "problems")=<externalptr>## [1] "alte" "bovine" "ovine"4 Subsetting
- Now that we know there are 3 livestock types (groups), subset the data to include only the rows for ‘ovine’
- In the same code chunk select the columns damage, year, month, bear_abund, landcover_code, and altitude
- Save this to your environment as ‘bear_sheep_data’
## # A tibble: 6 × 6
## damage year month bear_abund landcover_code altitude
## <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 1 2008 9 40 324 570
## 2 1 2008 9 40 324 570
## 3 1 2008 8 36 321 1410
## 4 1 2008 8 22 321 1068
## 5 1 2008 9 40 112 516
## 6 1 2008 8 56 112 5335 Summarise
- Using the bear_sheep_data and the
summarise()function, calculate the mean, sd and SE (the formula for SE is sd / sqrt(n)) for altitude. This last part might be tricky at first but give it a try and remember you can always google things and check the ‘help’ files; and if that fails ‘phone a friend or me’
## # A tibble: 1 × 3
## mean_alt sd_alt se_alt
## <dbl> <dbl> <dbl>
## 1 699. 183. 12.6Submission
Please email me a copy of your R script for this assignment by start of class on Friday February 2nd with your first and last name followed by assignment 2 as the file name (e.g. ‘marissa_dyck_assignment2.R’)
You should always be following best coding practices (see Intro to R module 1) but especially for assingment submissions. Please make sure each problem has its own header so that I can easily navigate to your answers and that your code is well organized with spaces as described in the best coding practices section and comments as needed.