Intro to R
Module 4: Data manipulation
9 & 16 February 2024
Materials
Script
Click here to download the script! Save the script to the ‘scripts’’ folder in your project directory you set up in the previous module.
Load your script in RStudio. To do this, open RStudio and click the files window and select the scripts folder and then this script.
Let’s get started formatting data!
Cheat sheets
tidyr and dplyr cheat sheets Scroll down to data tidying with tidyr & data transformation with dplyr
Import data
We will continue working with the turtles data set for part of this module, but let’s use the one we altered with lowercase and shorter variable names. Read in the altered turtles data frame we created in the last module (e.g. turtles_tidy) and print it to the console.
# import data FILL IN YOUR CODE HERE ----------------------
# read in altered turtles data
turtles_tidy <- read_csv('data/processed/turtles_tidy.csv')
# print data
turtles_tidy## # A tibble: 21 × 5
## tag sex c_length h_width weight
## <dbl> <chr> <dbl> <dbl> <dbl>
## 1 10 male 41 7.15 7.6
## 2 11 female 46.4 8.18 11
## 3 2 <NA> 24.3 4.42 1.65
## 4 15 <NA> 28.7 4.89 2.18
## 5 16 <NA> 32 5.37 3
## 6 3 female 42.8 7.32 8.6
## 7 4 male 40 6.6 6.5
## 8 5 female 45 8.05 10.9
## 9 12 female 44 7.55 8.9
## 10 13 <NA> 28 4.85 1.97
## # ℹ 11 more rows# check internal structure
str(turtles_tidy)## spc_tbl_ [21 × 5] (S3: spec_tbl_df/tbl_df/tbl/data.frame)
## $ tag : num [1:21] 10 11 2 15 16 3 4 5 12 13 ...
## $ sex : chr [1:21] "male" "female" NA NA ...
## $ c_length: num [1:21] 41 46.4 24.3 28.7 32 42.8 40 45 44 28 ...
## $ h_width : num [1:21] 7.15 8.18 4.42 4.89 5.37 7.32 6.6 8.05 7.55 4.85 ...
## $ weight : num [1:21] 7.6 11 1.65 2.18 3 8.6 6.5 10.9 8.9 1.97 ...
## - attr(*, "spec")=
## .. cols(
## .. tag = col_double(),
## .. sex = col_character(),
## .. c_length = col_double(),
## .. h_width = col_double(),
## .. weight = col_double()
## .. )
## - attr(*, "problems")=<externalptr>If you didn’t save the turtles_tidy data, below is the code to alter and save the data.
# code for altering turtle data
turtles.df <- read_delim('data/raw/turtle_data.txt',
delim = '\t') %>%
# set column names to lowercase
set_names(
names(.) %>%
tolower()) %>%
# rename columns to shorter names
rename(tag = tag_number,
c_length = carapace_length,
h_width = head_width) %>%
# change sex to factor variable
mutate(sex = as.factor(sex))
# save to data/processed folder as 'turtles_tidy.csv'
write_csv(turtles.df,
'data/processed/turtles_tidy.csv')Sorting/ordering data
Sorting data (rows)
Sorting is another common data operation, which helps to visualize
and organize data. In the tidyverse, sorting (by row)
is accomplished using the arrange()
function.
# Sorting/ordering data --------------------
# Sorting using arrange
# To sort a data frame by one vector (variable), you can use arrange()
turtles_tidy %>%
arrange(c_length)## # A tibble: 21 × 5
## tag sex c_length h_width weight
## <dbl> <chr> <dbl> <dbl> <dbl>
## 1 2 <NA> 24.3 4.42 1.65
## 2 13 <NA> 28 4.85 1.97
## 3 15 <NA> 28.7 4.89 2.18
## 4 1 <NA> 29.2 5.1 2.38
## 5 16 <NA> 32 5.37 3
## 6 8 <NA> 32 5.35 2.9
## 7 9 male 35 5.74 3.9
## 8 17 female 35.1 6.04 4.5
## 9 4 male 40 6.6 6.5
## 10 6 fem 40 6.53 6.2
## # ℹ 11 more rows# or if we want in descending order...
turtles_tidy %>%
arrange(desc(c_length))## # A tibble: 21 × 5
## tag sex c_length h_width weight
## <dbl> <chr> <dbl> <dbl> <dbl>
## 1 22 female 48.1 8.55 12.8
## 2 7 female 48 8.67 13.5
## 3 11 female 46.4 8.18 11
## 4 5 female 45 8.05 10.9
## 5 12 female 44 7.55 8.9
## 6 105 male 44 7.1 9
## 7 104 male 44 7.35 9
## 8 14 male 43 6.6 7.2
## 9 3 female 42.8 7.32 8.6
## 10 19 male 42.3 6.77 7.8
## # ℹ 11 more rows# Sorting by 2 columns - use a comma to separate columns
turtles_tidy %>%
arrange(sex, weight)## # A tibble: 21 × 5
## tag sex c_length h_width weight
## <dbl> <chr> <dbl> <dbl> <dbl>
## 1 6 fem 40 6.53 6.2
## 2 17 female 35.1 6.04 4.5
## 3 3 female 42.8 7.32 8.6
## 4 12 female 44 7.55 8.9
## 5 5 female 45 8.05 10.9
## 6 11 female 46.4 8.18 11
## 7 22 female 48.1 8.55 12.8
## 8 7 female 48 8.67 13.5
## 9 9 male 35 5.74 3.9
## 10 4 male 40 6.6 6.5
## # ℹ 11 more rowsLet’s practice this with the diamonds data in the ggplot2 package. Arrange the data by highest price to lowest price.
# Practice FILL IN YOUR CODE HERE ----------------------
# practice- arrange diamonds data by highest price to lowest priceOrdering data (columns)
We can also reorder the columns in our data set using
relocate() or select().
Select
In the turtles_tidy data set if we want the columns ordered
alphabetically we can use the select()
function paired with the order()
function
# order columns alphabetically with select
turtles_tidy %>%
select(order(colnames(.))) ## # A tibble: 21 × 5
## c_length h_width sex tag weight
## <dbl> <dbl> <chr> <dbl> <dbl>
## 1 41 7.15 male 10 7.6
## 2 46.4 8.18 female 11 11
## 3 24.3 4.42 <NA> 2 1.65
## 4 28.7 4.89 <NA> 15 2.18
## 5 32 5.37 <NA> 16 3
## 6 42.8 7.32 female 3 8.6
## 7 40 6.6 male 4 6.5
## 8 45 8.05 female 5 10.9
## 9 44 7.55 female 12 8.9
## 10 28 4.85 <NA> 13 1.97
## # ℹ 11 more rowsHere we use dplyr select() to tell R we want to do
something with the columns in our data. Then we specify we want to order
the columns using order(), then inside that
function we use colnames() to specify that
we want to order the columns based on their names. The ‘.’ inside the
colnames() function is a placeholder for
the data (e.g. turtles_tidy) which we are piping in above. If we weren’t
using a dplyr pipe we would need to specify the data inside
colnames().
Relocate
We can also specify the location of specific columns relative to
other columns in our data using relocate().
In the turtles_tidy data if we want ‘weight’ to come before ‘c_length’ we can reorder the columns as follows
# ordering columns with relocate
# example with turtles_tidy data
turtles_tidy %>%
# move 'weight' before 'c_length'
relocate(weight,
.before = c_length)## # A tibble: 21 × 5
## tag sex weight c_length h_width
## <dbl> <chr> <dbl> <dbl> <dbl>
## 1 10 male 7.6 41 7.15
## 2 11 female 11 46.4 8.18
## 3 2 <NA> 1.65 24.3 4.42
## 4 15 <NA> 2.18 28.7 4.89
## 5 16 <NA> 3 32 5.37
## 6 3 female 8.6 42.8 7.32
## 7 4 male 6.5 40 6.6
## 8 5 female 10.9 45 8.05
## 9 12 female 8.9 44 7.55
## 10 13 <NA> 1.97 28 4.85
## # ℹ 11 more rows# alternatively we could specify to have weight AFTER 'sex'
turtles_tidy %>%
# relocate 'weight' after 'sex'
relocate(weight,
.after = sex)## # A tibble: 21 × 5
## tag sex weight c_length h_width
## <dbl> <chr> <dbl> <dbl> <dbl>
## 1 10 male 7.6 41 7.15
## 2 11 female 11 46.4 8.18
## 3 2 <NA> 1.65 24.3 4.42
## 4 15 <NA> 2.18 28.7 4.89
## 5 16 <NA> 3 32 5.37
## 6 3 female 8.6 42.8 7.32
## 7 4 male 6.5 40 6.6
## 8 5 female 10.9 45 8.05
## 9 12 female 8.9 44 7.55
## 10 13 <NA> 1.97 28 4.85
## # ℹ 11 more rows# if we don't specify a .before or .after argument it will move the selected column to the front of the data
turtles_tidy %>%
relocate(weight)## # A tibble: 21 × 5
## weight tag sex c_length h_width
## <dbl> <dbl> <chr> <dbl> <dbl>
## 1 7.6 10 male 41 7.15
## 2 11 11 female 46.4 8.18
## 3 1.65 2 <NA> 24.3 4.42
## 4 2.18 15 <NA> 28.7 4.89
## 5 3 16 <NA> 32 5.37
## 6 8.6 3 female 42.8 7.32
## 7 6.5 4 male 40 6.6
## 8 10.9 5 female 45 8.05
## 9 8.9 12 female 44 7.55
## 10 1.97 13 <NA> 28 4.85
## # ℹ 11 more rowsRemember unless we overwrite the data (e.g. assign it to the environment with
<-) none of these changes will be saved outside the code chunk we wrote them in.
We can also reorder multiple columns at a time
# ordering data
# reordering multiple columns by name
turtles_tidy %>%
# move 'weight', 'c_length', and 'h_width' before 'tag' and 'sex'
relocate(c(weight, c_length, h_width))## # A tibble: 21 × 5
## weight c_length h_width tag sex
## <dbl> <dbl> <dbl> <dbl> <chr>
## 1 7.6 41 7.15 10 male
## 2 11 46.4 8.18 11 female
## 3 1.65 24.3 4.42 2 <NA>
## 4 2.18 28.7 4.89 15 <NA>
## 5 3 32 5.37 16 <NA>
## 6 8.6 42.8 7.32 3 female
## 7 6.5 40 6.6 4 male
## 8 10.9 45 8.05 5 female
## 9 8.9 44 7.55 12 female
## 10 1.97 28 4.85 13 <NA>
## # ℹ 11 more rows# alternatively we could specify which variables to relocate by their type
turtles_tidy %>%
# move all numeric variables to the front
relocate(where(is.numeric))## # A tibble: 21 × 5
## tag c_length h_width weight sex
## <dbl> <dbl> <dbl> <dbl> <chr>
## 1 10 41 7.15 7.6 male
## 2 11 46.4 8.18 11 female
## 3 2 24.3 4.42 1.65 <NA>
## 4 15 28.7 4.89 2.18 <NA>
## 5 16 32 5.37 3 <NA>
## 6 3 42.8 7.32 8.6 female
## 7 4 40 6.6 6.5 male
## 8 5 45 8.05 10.9 female
## 9 12 44 7.55 8.9 female
## 10 13 28 4.85 1.97 <NA>
## # ℹ 11 more rows# or
turtles_tidy %>%
# move all numeric variables after 'tag'
relocate(where(is.numeric),
.after = tag)## # A tibble: 21 × 5
## tag c_length h_width weight sex
## <dbl> <dbl> <dbl> <dbl> <chr>
## 1 10 41 7.15 7.6 male
## 2 11 46.4 8.18 11 female
## 3 2 24.3 4.42 1.65 <NA>
## 4 15 28.7 4.89 2.18 <NA>
## 5 16 32 5.37 3 <NA>
## 6 3 42.8 7.32 8.6 female
## 7 4 40 6.6 6.5 male
## 8 5 45 8.05 10.9 female
## 9 12 44 7.55 8.9 female
## 10 13 28 4.85 1.97 <NA>
## # ℹ 11 more rowsPractice with the diamonds data in the ggplot2 package. Reorder the columns so that price is first.
# practice with diamonds data
# reorder columns so price is first
diamonds %>%
# move price to front
relocate(price) # most parsimonious way## # A tibble: 53,940 × 10
## price carat cut color clarity depth table x y z
## <int> <dbl> <ord> <ord> <ord> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 326 0.23 Ideal E SI2 61.5 55 3.95 3.98 2.43
## 2 326 0.21 Premium E SI1 59.8 61 3.89 3.84 2.31
## 3 327 0.23 Good E VS1 56.9 65 4.05 4.07 2.31
## 4 334 0.29 Premium I VS2 62.4 58 4.2 4.23 2.63
## 5 335 0.31 Good J SI2 63.3 58 4.34 4.35 2.75
## 6 336 0.24 Very Good J VVS2 62.8 57 3.94 3.96 2.48
## 7 336 0.24 Very Good I VVS1 62.3 57 3.95 3.98 2.47
## 8 337 0.26 Very Good H SI1 61.9 55 4.07 4.11 2.53
## 9 337 0.22 Fair E VS2 65.1 61 3.87 3.78 2.49
## 10 338 0.23 Very Good H VS1 59.4 61 4 4.05 2.39
## # ℹ 53,930 more rows# or
diamonds %>%
# move price to front
relocate(price,
.before = carat)## # A tibble: 53,940 × 10
## price carat cut color clarity depth table x y z
## <int> <dbl> <ord> <ord> <ord> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 326 0.23 Ideal E SI2 61.5 55 3.95 3.98 2.43
## 2 326 0.21 Premium E SI1 59.8 61 3.89 3.84 2.31
## 3 327 0.23 Good E VS1 56.9 65 4.05 4.07 2.31
## 4 334 0.29 Premium I VS2 62.4 58 4.2 4.23 2.63
## 5 335 0.31 Good J SI2 63.3 58 4.34 4.35 2.75
## 6 336 0.24 Very Good J VVS2 62.8 57 3.94 3.96 2.48
## 7 336 0.24 Very Good I VVS1 62.3 57 3.95 3.98 2.47
## 8 337 0.26 Very Good H SI1 61.9 55 4.07 4.11 2.53
## 9 337 0.22 Fair E VS2 65.1 61 3.87 3.78 2.49
## 10 338 0.23 Very Good H VS1 59.4 61 4 4.05 2.39
## # ℹ 53,930 more rowsWith the turtles_tidy data let’s try to move all the factors to the front
turtles_tidy %>%
# move factors to front
relocate(where(is.factor))## # A tibble: 21 × 5
## tag sex c_length h_width weight
## <dbl> <chr> <dbl> <dbl> <dbl>
## 1 10 male 41 7.15 7.6
## 2 11 female 46.4 8.18 11
## 3 2 <NA> 24.3 4.42 1.65
## 4 15 <NA> 28.7 4.89 2.18
## 5 16 <NA> 32 5.37 3
## 6 3 female 42.8 7.32 8.6
## 7 4 male 40 6.6 6.5
## 8 5 female 45 8.05 10.9
## 9 12 female 44 7.55 8.9
## 10 13 <NA> 28 4.85 1.97
## # ℹ 11 more rowsNotice this didn’t change our data… It should have moved ‘sex’ to the front because we set it as a factor last time. Let’s check the data structure again to be sure.
# check structure of turtles_tidy
str(turtles_tidy)## spc_tbl_ [21 × 5] (S3: spec_tbl_df/tbl_df/tbl/data.frame)
## $ tag : num [1:21] 10 11 2 15 16 3 4 5 12 13 ...
## $ sex : chr [1:21] "male" "female" NA NA ...
## $ c_length: num [1:21] 41 46.4 24.3 28.7 32 42.8 40 45 44 28 ...
## $ h_width : num [1:21] 7.15 8.18 4.42 4.89 5.37 7.32 6.6 8.05 7.55 4.85 ...
## $ weight : num [1:21] 7.6 11 1.65 2.18 3 8.6 6.5 10.9 8.9 1.97 ...
## - attr(*, "spec")=
## .. cols(
## .. tag = col_double(),
## .. sex = col_character(),
## .. c_length = col_double(),
## .. h_width = col_double(),
## .. weight = col_double()
## .. )
## - attr(*, "problems")=<externalptr>It is still reading ‘sex’ as a character. Sometimes R will do this but luckily you know how to change the type of variable when you read in the data.
Data manipulation
Alter the code we used to read in the turtles_tidy data to change sex to a factor the same code chunk and then check the internal data structure again..
# Data manipulation --------------------
# FILL IN YOUR CODE HERE --------------------
# first read in data for this section
# read in altered turtles data
turtles_tidy <- read_csv('data/processed/turtles_tidy.csv') %>%
# change 'sex' to a factor
mutate(sex = as.factor(sex))
# check internal structure
str(turtles_tidy)## tibble [21 × 5] (S3: tbl_df/tbl/data.frame)
## $ tag : num [1:21] 10 11 2 15 16 3 4 5 12 13 ...
## $ sex : Factor w/ 3 levels "fem","female",..: 3 2 NA NA NA 2 3 2 2 NA ...
## $ c_length: num [1:21] 41 46.4 24.3 28.7 32 42.8 40 45 44 28 ...
## $ h_width : num [1:21] 7.15 8.18 4.42 4.89 5.37 7.32 6.6 8.05 7.55 4.85 ...
## $ weight : num [1:21] 7.6 11 1.65 2.18 3 8.6 6.5 10.9 8.9 1.97 ...Do you notice anything else that could be a problem with the sex variable in this data set? Maybe you noticed this in the last module already. Females has been entered two different ways (‘female’ and ‘fem’). This is a very common problem that can arise with data, particularly when multiple people are in charge of entering the data. There are multiple ways to fix this, let’s look at a few using tidyverse.
Replace
First, we can use the mutate() and
replace() functions. Recall that
mutate() can create new columns or modify existing columns
using existing variables in the data. Therefore, we can modify the sex
column using this function and another
function, replace() which as the name
suggests will replace existing values matching some criteria with a
specified value.
# Replace ---------------------
# fix issue with multiple entries for female
turtles_tidy <- turtles_tidy %>%
mutate(sex = replace(sex,
sex == "fem",
"female"))
# print
turtles_tidy## # A tibble: 21 × 5
## tag sex c_length h_width weight
## <dbl> <fct> <dbl> <dbl> <dbl>
## 1 10 male 41 7.15 7.6
## 2 11 female 46.4 8.18 11
## 3 2 <NA> 24.3 4.42 1.65
## 4 15 <NA> 28.7 4.89 2.18
## 5 16 <NA> 32 5.37 3
## 6 3 female 42.8 7.32 8.6
## 7 4 male 40 6.6 6.5
## 8 5 female 45 8.05 10.9
## 9 12 female 44 7.55 8.9
## 10 13 <NA> 28 4.85 1.97
## # ℹ 11 more rowslevels(turtles_tidy$sex)## [1] "fem" "female" "male"Now notice this replaced ‘fem’ with ‘female’ when we print the data but when we look at the levels of our variable ‘sex’ it still shows 3 levels (‘fem’, ‘female’, ‘male’). It took me a little while to figure out why this wasn’t working, do you have any ideas?

I thought it might have to do with the fact that our variable is a factor. How could I test this to see if this is in fact the issue?
# FILL IN YOUR CODE HERE --------------------
# change 'sex' back to a character then try the data manipulation again to see if anything is different. We can do this all in one pipe!
turtles_tidy <- turtles_tidy %>%
# sex to character
mutate(sex = as.character(sex),
# change fem to female with replace
replace(sex,
sex == 'fem',
'female'),
# sex to factor
sex = as.factor(sex))
head(turtles_tidy)## # A tibble: 6 × 6
## tag sex c_length h_width weight `replace(sex, sex == "fem", "female")`
## <dbl> <fct> <dbl> <dbl> <dbl> <chr>
## 1 10 male 41 7.15 7.6 male
## 2 11 female 46.4 8.18 11 female
## 3 2 <NA> 24.3 4.42 1.65 <NA>
## 4 15 <NA> 28.7 4.89 2.18 <NA>
## 5 16 <NA> 32 5.37 3 <NA>
## 6 3 female 42.8 7.32 8.6 femalelevels(turtles_tidy$sex)## [1] "female" "male"Recode
It turns out we were correct, replace() which is a base
R function wasn’t working properly with a
factor. Luckily, dplyr has a similar
function to ‘replace’ values which works better with
factors, it is recode() and here is how it
works.
# Recode ----------------------
# read in turtles data again and set 'sex' to factor to overwrite the changes we made with the last code chunk, and in the same code chunk we will 'recode' the 'sex' column
# read in altered turtles data
turtles_tidy <- read_csv('data/processed/turtles_tidy.csv') %>%
# change sex to a factor
mutate(sex = as.factor(sex),
sex = recode(sex,
fem = 'female'))
# check data
str(turtles_tidy)## tibble [21 × 5] (S3: tbl_df/tbl/data.frame)
## $ tag : num [1:21] 10 11 2 15 16 3 4 5 12 13 ...
## $ sex : Factor w/ 2 levels "female","male": 2 1 NA NA NA 1 2 1 1 NA ...
## $ c_length: num [1:21] 41 46.4 24.3 28.7 32 42.8 40 45 44 28 ...
## $ h_width : num [1:21] 7.15 8.18 4.42 4.89 5.37 7.32 6.6 8.05 7.55 4.85 ...
## $ weight : num [1:21] 7.6 11 1.65 2.18 3 8.6 6.5 10.9 8.9 1.97 ...levels(turtles_tidy$sex)## [1] "female" "male"Now you might wonder why I bothered going through the trouble of
first showing you replace() when it didn’t work and the
lengthy way to fix it above.
This is because all data are different and you will find when working with your own data that code which works perfectly in an example, online, or in other code you have run isn’t working. And rather than get frustrated or give up and try a different function or package or even worse…. try to manually fix it in… EXCEL! You will need to learn to troubleshoot your code, and this provides a great example of that.

Rant over, let’s move on to other ways we can correct/modify entries in a column.
If_else
Another useful function is if_else().
If-else statements are logical statements that can alter or replace
existing data based on whether the value in a column matches your
criteria (TRUE), does not match criteria (FALSE), or is missing
data.
Let’s use the diamonds data set in the ggplot2
package for this example. We can use the help shortcut
? to learn more about the data. By reading this we see that
color (D-J) is a ranking from best to worst. Maybe we don’t want 7
levels for this variable though, if we just want to know which ones are
the worst color and which ones are not we can use
if_else().
# If else ----------------------
# first learn morn about the diamonds data set
?diamonds
# change color to two categories based on worst color diamonds (J)
diamonds %>%
mutate(color = if_else(color == 'J', # if color is J
'worst', # TRUE = worst
'not_worst')) # FALSE = not_worst## # A tibble: 53,940 × 10
## carat cut color clarity depth table price x y z
## <dbl> <ord> <chr> <ord> <dbl> <dbl> <int> <dbl> <dbl> <dbl>
## 1 0.23 Ideal not_worst SI2 61.5 55 326 3.95 3.98 2.43
## 2 0.21 Premium not_worst SI1 59.8 61 326 3.89 3.84 2.31
## 3 0.23 Good not_worst VS1 56.9 65 327 4.05 4.07 2.31
## 4 0.29 Premium not_worst VS2 62.4 58 334 4.2 4.23 2.63
## 5 0.31 Good worst SI2 63.3 58 335 4.34 4.35 2.75
## 6 0.24 Very Good worst VVS2 62.8 57 336 3.94 3.96 2.48
## 7 0.24 Very Good not_worst VVS1 62.3 57 336 3.95 3.98 2.47
## 8 0.26 Very Good not_worst SI1 61.9 55 337 4.07 4.11 2.53
## 9 0.22 Fair not_worst VS2 65.1 61 337 3.87 3.78 2.49
## 10 0.23 Very Good not_worst VS1 59.4 61 338 4 4.05 2.39
## # ℹ 53,930 more rowsWe could also create a new column for this variable instead if we want to preserve the original data
# create new column with two categories based on worst color diamonds (J)
diamonds %>%
mutate(color_quality = if_else(color == 'J', # if color is J
'worst', # TRUE = worst
'not_worst')) # FALSE = not_worst## # A tibble: 53,940 × 11
## carat cut color clarity depth table price x y z color_quality
## <dbl> <ord> <ord> <ord> <dbl> <dbl> <int> <dbl> <dbl> <dbl> <chr>
## 1 0.23 Ideal E SI2 61.5 55 326 3.95 3.98 2.43 not_worst
## 2 0.21 Premium E SI1 59.8 61 326 3.89 3.84 2.31 not_worst
## 3 0.23 Good E VS1 56.9 65 327 4.05 4.07 2.31 not_worst
## 4 0.29 Premium I VS2 62.4 58 334 4.2 4.23 2.63 not_worst
## 5 0.31 Good J SI2 63.3 58 335 4.34 4.35 2.75 worst
## 6 0.24 Very G… J VVS2 62.8 57 336 3.94 3.96 2.48 worst
## 7 0.24 Very G… I VVS1 62.3 57 336 3.95 3.98 2.47 not_worst
## 8 0.26 Very G… H SI1 61.9 55 337 4.07 4.11 2.53 not_worst
## 9 0.22 Fair E VS2 65.1 61 337 3.87 3.78 2.49 not_worst
## 10 0.23 Very G… H VS1 59.4 61 338 4 4.05 2.39 not_worst
## # ℹ 53,930 more rowsCase_when
A useful variation of if_else() is
case_when() which allows you to use multiple if-else
statements to modify data. For example, the clarity column in the
diamonds data set isn’t easy to interpret if you aren’t familiar with
this measurement so we can change the names to something easier to
understand
# Case when ----------------------
# create new column with two categories based on worst color diamonds (J)
diamonds %>%
mutate(clarity = case_when(clarity == 'l1' ~ 8,
clarity == 'SI2' ~ 7,
clarity == 'SI1' ~ 6,
clarity == 'VS2' ~ 5,
clarity == 'VS1' ~ 4,
clarity == 'VVS2' ~ 3,
clarity == 'VVS1' ~ 2,
clarity == 'IF' ~ 1))## # A tibble: 53,940 × 10
## carat cut color clarity depth table price x y z
## <dbl> <ord> <ord> <dbl> <dbl> <dbl> <int> <dbl> <dbl> <dbl>
## 1 0.23 Ideal E 7 61.5 55 326 3.95 3.98 2.43
## 2 0.21 Premium E 6 59.8 61 326 3.89 3.84 2.31
## 3 0.23 Good E 4 56.9 65 327 4.05 4.07 2.31
## 4 0.29 Premium I 5 62.4 58 334 4.2 4.23 2.63
## 5 0.31 Good J 7 63.3 58 335 4.34 4.35 2.75
## 6 0.24 Very Good J 3 62.8 57 336 3.94 3.96 2.48
## 7 0.24 Very Good I 2 62.3 57 336 3.95 3.98 2.47
## 8 0.26 Very Good H 6 61.9 55 337 4.07 4.11 2.53
## 9 0.22 Fair E 5 65.1 61 337 3.87 3.78 2.49
## 10 0.23 Very Good H 4 59.4 61 338 4 4.05 2.39
## # ℹ 53,930 more rowsThe syntax for case_when() can seem confusing at first,
but as with other tidyverse operations, code can be read left
to right. Inside the case_when() function,
first reference the variable name (e.g. clarity) followed by
the boolean operation (e.g. equals, greater than, less
than, etc.), then the entry you are performing the operation on
(e.g. l1, SI2, etc.) and finally a ‘~’ followed by the new entry you
want.
Now we have scored the diamonds based on clarity with 8 being the worst and 1 being the best.
This is a lot of categories/levels though. Maybe if we were doing an analysis or plotting this data we would want to simplify this data and clump some of these scores together. We can do that too!
# group the scores for clarity using case_when
diamonds %>%
mutate(clarity = case_when(clarity == 'l1' ~ 'worst',
clarity == 'IF' ~ 'best',
.default = 'neutral'))## # A tibble: 53,940 × 10
## carat cut color clarity depth table price x y z
## <dbl> <ord> <ord> <chr> <dbl> <dbl> <int> <dbl> <dbl> <dbl>
## 1 0.23 Ideal E neutral 61.5 55 326 3.95 3.98 2.43
## 2 0.21 Premium E neutral 59.8 61 326 3.89 3.84 2.31
## 3 0.23 Good E neutral 56.9 65 327 4.05 4.07 2.31
## 4 0.29 Premium I neutral 62.4 58 334 4.2 4.23 2.63
## 5 0.31 Good J neutral 63.3 58 335 4.34 4.35 2.75
## 6 0.24 Very Good J neutral 62.8 57 336 3.94 3.96 2.48
## 7 0.24 Very Good I neutral 62.3 57 336 3.95 3.98 2.47
## 8 0.26 Very Good H neutral 61.9 55 337 4.07 4.11 2.53
## 9 0.22 Fair E neutral 65.1 61 337 3.87 3.78 2.49
## 10 0.23 Very Good H neutral 59.4 61 338 4 4.05 2.39
## # ℹ 53,930 more rowsWhat we did here is define the worst and best scores for clarity and
then using the .default argument we set everything else as
‘neutral’.
We can also set multiple values as the same new value.
# group the scores for clarity using case_when
diamonds %>%
mutate(clarity = case_when(clarity == 'l1' ~ 'poor',
clarity == 'SI2' ~ 'poor',
clarity == 'WS1' ~ 'good',
clarity == 'IF' ~ 'good',
.default = 'moderate'))## # A tibble: 53,940 × 10
## carat cut color clarity depth table price x y z
## <dbl> <ord> <ord> <chr> <dbl> <dbl> <int> <dbl> <dbl> <dbl>
## 1 0.23 Ideal E poor 61.5 55 326 3.95 3.98 2.43
## 2 0.21 Premium E moderate 59.8 61 326 3.89 3.84 2.31
## 3 0.23 Good E moderate 56.9 65 327 4.05 4.07 2.31
## 4 0.29 Premium I moderate 62.4 58 334 4.2 4.23 2.63
## 5 0.31 Good J poor 63.3 58 335 4.34 4.35 2.75
## 6 0.24 Very Good J moderate 62.8 57 336 3.94 3.96 2.48
## 7 0.24 Very Good I moderate 62.3 57 336 3.95 3.98 2.47
## 8 0.26 Very Good H moderate 61.9 55 337 4.07 4.11 2.53
## 9 0.22 Fair E moderate 65.1 61 337 3.87 3.78 2.49
## 10 0.23 Very Good H moderate 59.4 61 338 4 4.05 2.39
## # ℹ 53,930 more rowsBut, if we have a lot of different values we want to change to the
same new value we can also define a vector with those first and then use
case_when() to overwrite them. For example.
# using a vector with case_when
# define the entries we want to be 'poor' quality
poor_quality <- c('l1', 'SI2')
# define the entries we want to be 'good' quality
good_quality <- c('WS1', 'IF')
# group the scores for clarity using case_when and %in%
diamonds %>%
mutate(clarity = case_when(clarity %in% poor_quality ~ 'poor',
clarity %in% good_quality ~ 'good',
.default = 'moderate'))## # A tibble: 53,940 × 10
## carat cut color clarity depth table price x y z
## <dbl> <ord> <ord> <chr> <dbl> <dbl> <int> <dbl> <dbl> <dbl>
## 1 0.23 Ideal E poor 61.5 55 326 3.95 3.98 2.43
## 2 0.21 Premium E moderate 59.8 61 326 3.89 3.84 2.31
## 3 0.23 Good E moderate 56.9 65 327 4.05 4.07 2.31
## 4 0.29 Premium I moderate 62.4 58 334 4.2 4.23 2.63
## 5 0.31 Good J poor 63.3 58 335 4.34 4.35 2.75
## 6 0.24 Very Good J moderate 62.8 57 336 3.94 3.96 2.48
## 7 0.24 Very Good I moderate 62.3 57 336 3.95 3.98 2.47
## 8 0.26 Very Good H moderate 61.9 55 337 4.07 4.11 2.53
## 9 0.22 Fair E moderate 65.1 61 337 3.87 3.78 2.49
## 10 0.23 Very Good H moderate 59.4 61 338 4 4.05 2.39
## # ℹ 53,930 more rowsFinally we can also combine other boolean operations
with mutate() to modify data. For example we could change
price from a numerical variable to a categorical one based on the value
in that column
# using other boolean operations with mutate()
# look at range for price
summary(diamonds$price) # let's use the mean and 1st and 3rd quartiles to set our levels## Min. 1st Qu. Median Mean 3rd Qu. Max.
## 326 950 2401 3933 5324 18823# change price to factor/categorical
diamonds %>%
mutate(price = case_when(price < 950 ~ 'low',
price > 950 & price < 5324 ~ 'moderate',
price > 5324 ~ 'high'))## # A tibble: 53,940 × 10
## carat cut color clarity depth table price x y z
## <dbl> <ord> <ord> <ord> <dbl> <dbl> <chr> <dbl> <dbl> <dbl>
## 1 0.23 Ideal E SI2 61.5 55 low 3.95 3.98 2.43
## 2 0.21 Premium E SI1 59.8 61 low 3.89 3.84 2.31
## 3 0.23 Good E VS1 56.9 65 low 4.05 4.07 2.31
## 4 0.29 Premium I VS2 62.4 58 low 4.2 4.23 2.63
## 5 0.31 Good J SI2 63.3 58 low 4.34 4.35 2.75
## 6 0.24 Very Good J VVS2 62.8 57 low 3.94 3.96 2.48
## 7 0.24 Very Good I VVS1 62.3 57 low 3.95 3.98 2.47
## 8 0.26 Very Good H SI1 61.9 55 low 4.07 4.11 2.53
## 9 0.22 Fair E VS2 65.1 61 low 3.87 3.78 2.49
## 10 0.23 Very Good H VS1 59.4 61 low 4 4.05 2.39
## # ℹ 53,930 more rows# or using the default argument
diamonds %>%
mutate(price = case_when(price < 950 ~ 'low',
price > 5324 ~ 'high',
.default = 'moderate'))## # A tibble: 53,940 × 10
## carat cut color clarity depth table price x y z
## <dbl> <ord> <ord> <ord> <dbl> <dbl> <chr> <dbl> <dbl> <dbl>
## 1 0.23 Ideal E SI2 61.5 55 low 3.95 3.98 2.43
## 2 0.21 Premium E SI1 59.8 61 low 3.89 3.84 2.31
## 3 0.23 Good E VS1 56.9 65 low 4.05 4.07 2.31
## 4 0.29 Premium I VS2 62.4 58 low 4.2 4.23 2.63
## 5 0.31 Good J SI2 63.3 58 low 4.34 4.35 2.75
## 6 0.24 Very Good J VVS2 62.8 57 low 3.94 3.96 2.48
## 7 0.24 Very Good I VVS1 62.3 57 low 3.95 3.98 2.47
## 8 0.26 Very Good H SI1 61.9 55 low 4.07 4.11 2.53
## 9 0.22 Fair E VS2 65.1 61 low 3.87 3.78 2.49
## 10 0.23 Very Good H VS1 59.4 61 low 4 4.05 2.39
## # ℹ 53,930 more rowsAs you can see there are a lot of ways to modify data using
mutate(), and these are just a few examples to get you
started.
Be very careful when using the .default agrument. It will alter the value for any remaining rows including NAs.
Dealing w/ NAs in data
Speaking of NAs, in many real-world data sets, observations are incomplete in some way- they are missing information.
In R, the code “NA” (not available) stands in for elements of a vector that are missing for whatever reason. Most statistical functions have ways of dealing with NAs, but you may want to modify your data yourself to deal with NAs.
Let’s explore a data set with missing data.
If you haven’t already, download the Example missing data (data_missing.txt) and save to the data folder in your project directory.
Check for NAs
Now let’s explore this data set in more detail, and go over a few ways to check for NAs
# Dealing with NAs ----------------------
# Check for NAs ----------------------
# read data
# NOTE: you need to specify that this is a tab-delimited file. It is especially important to specify the delimiter for data files with missing data. If you specify the header and what the text is delimited by correctly, it will read missing data as NA. Otherwise it will fail to read data in properly.
missing.df <- read_delim('data/raw/data_missing.txt',
delim = "\t") %>%
# set names to lowercase
set_names(
names(.) %>% # the period here is a placeholder for the data it tells R to use the element before the last pipe (e.g., turtles.df)
tolower())
# Missing data are read as an NA
missing.df## # A tibble: 20 × 4
## country import export product
## <chr> <dbl> <dbl> <chr>
## 1 Bolivia 46 0 N
## 2 Brazil 74 0 N
## 3 Chile 89 16 N
## 4 Colombia 77 16 A
## 5 CostaRica 84 21 A
## 6 Cuba 89 15 A
## 7 DominicanRep NA 14 A
## 8 Ecuador 70 6 A
## 9 ElSalvador 60 13 A
## 10 Guatemala 55 9 A
## 11 Haiti 35 3 A
## 12 Honduras 51 NA A
## 13 Jamaica 87 23 A
## 14 Mexico 83 4 N
## 15 Nicaragua 68 0 A
## 16 Panama NA 19 N
## 17 Paraguay 74 3 A
## 18 Peru 73 NA N
## 19 TrinidadTobago 84 15 A
## 20 Venezuela NA 7 N# You can summarize your data and tell you how many NA's per col
summary(missing.df)## country import export product
## Length:20 Min. :35.00 Min. : 0.00 Length:20
## Class :character 1st Qu.:60.00 1st Qu.: 3.25 Class :character
## Mode :character Median :74.00 Median :11.00 Mode :character
## Mean :70.53 Mean :10.22
## 3rd Qu.:84.00 3rd Qu.:15.75
## Max. :89.00 Max. :23.00
## NA's :3 NA's :2# ?is.na (Boolean test!)
is.na(missing.df)## country import export product
## [1,] FALSE FALSE FALSE FALSE
## [2,] FALSE FALSE FALSE FALSE
## [3,] FALSE FALSE FALSE FALSE
## [4,] FALSE FALSE FALSE FALSE
## [5,] FALSE FALSE FALSE FALSE
## [6,] FALSE FALSE FALSE FALSE
## [7,] FALSE TRUE FALSE FALSE
## [8,] FALSE FALSE FALSE FALSE
## [9,] FALSE FALSE FALSE FALSE
## [10,] FALSE FALSE FALSE FALSE
## [11,] FALSE FALSE FALSE FALSE
## [12,] FALSE FALSE TRUE FALSE
## [13,] FALSE FALSE FALSE FALSE
## [14,] FALSE FALSE FALSE FALSE
## [15,] FALSE FALSE FALSE FALSE
## [16,] FALSE TRUE FALSE FALSE
## [17,] FALSE FALSE FALSE FALSE
## [18,] FALSE FALSE TRUE FALSE
## [19,] FALSE FALSE FALSE FALSE
## [20,] FALSE TRUE FALSE FALSEcomplete.cases(missing.df) # Boolean: for each row, tests if there are no NA values## [1] TRUE TRUE TRUE TRUE TRUE TRUE FALSE TRUE TRUE TRUE TRUE FALSE
## [13] TRUE TRUE TRUE FALSE TRUE FALSE TRUE FALSERemoving rows with NAs
# Remove NAs ----------------------
# Base R function that omits (removes) rows with missing data
na.omit(missing.df) # note this removes the entire row, so only do this if you don't want to use any data from an observation with NAs## # A tibble: 15 × 4
## country import export product
## <chr> <dbl> <dbl> <chr>
## 1 Bolivia 46 0 N
## 2 Brazil 74 0 N
## 3 Chile 89 16 N
## 4 Colombia 77 16 A
## 5 CostaRica 84 21 A
## 6 Cuba 89 15 A
## 7 Ecuador 70 6 A
## 8 ElSalvador 60 13 A
## 9 Guatemala 55 9 A
## 10 Haiti 35 3 A
## 11 Jamaica 87 23 A
## 12 Mexico 83 4 N
## 13 Nicaragua 68 0 A
## 14 Paraguay 74 3 A
## 15 TrinidadTobago 84 15 A# we can also use this with %>% to remove rows with NA
missing.df %>%
# remove rows with NA
na.omit()## # A tibble: 15 × 4
## country import export product
## <chr> <dbl> <dbl> <chr>
## 1 Bolivia 46 0 N
## 2 Brazil 74 0 N
## 3 Chile 89 16 N
## 4 Colombia 77 16 A
## 5 CostaRica 84 21 A
## 6 Cuba 89 15 A
## 7 Ecuador 70 6 A
## 8 ElSalvador 60 13 A
## 9 Guatemala 55 9 A
## 10 Haiti 35 3 A
## 11 Jamaica 87 23 A
## 12 Mexico 83 4 N
## 13 Nicaragua 68 0 A
## 14 Paraguay 74 3 A
## 15 TrinidadTobago 84 15 AYou may also want to remove only rows that contain NAs for a
specified column. Let’s say it doesn’t matter if an observation is
missing data for variables X and Y but if there is an NA for variable Z
then we can’t use that observation. We can do this using the
drop_na() function.
# print missing.df data to compare with data after we remove NAs
missing.df## # A tibble: 20 × 4
## country import export product
## <chr> <dbl> <dbl> <chr>
## 1 Bolivia 46 0 N
## 2 Brazil 74 0 N
## 3 Chile 89 16 N
## 4 Colombia 77 16 A
## 5 CostaRica 84 21 A
## 6 Cuba 89 15 A
## 7 DominicanRep NA 14 A
## 8 Ecuador 70 6 A
## 9 ElSalvador 60 13 A
## 10 Guatemala 55 9 A
## 11 Haiti 35 3 A
## 12 Honduras 51 NA A
## 13 Jamaica 87 23 A
## 14 Mexico 83 4 N
## 15 Nicaragua 68 0 A
## 16 Panama NA 19 N
## 17 Paraguay 74 3 A
## 18 Peru 73 NA N
## 19 TrinidadTobago 84 15 A
## 20 Venezuela NA 7 N# use drop_na to remove only rows missing data for 'import'
missing.df %>%
# remove NAs
drop_na(import)## # A tibble: 17 × 4
## country import export product
## <chr> <dbl> <dbl> <chr>
## 1 Bolivia 46 0 N
## 2 Brazil 74 0 N
## 3 Chile 89 16 N
## 4 Colombia 77 16 A
## 5 CostaRica 84 21 A
## 6 Cuba 89 15 A
## 7 Ecuador 70 6 A
## 8 ElSalvador 60 13 A
## 9 Guatemala 55 9 A
## 10 Haiti 35 3 A
## 11 Honduras 51 NA A
## 12 Jamaica 87 23 A
## 13 Mexico 83 4 N
## 14 Nicaragua 68 0 A
## 15 Paraguay 74 3 A
## 16 Peru 73 NA N
## 17 TrinidadTobago 84 15 AWe can also do this for multiple columns.
Below we use drop_na to remove only rows missing data for ‘import’ and ‘export’ (this is effectively the same as using na.omit for this data set because those are the only columns with NAs but it shows you how to drop rows for multiple variables)
# drop NAs for import and export columns
missing.df %>%
# remove NAs
drop_na(c(import,
export))## # A tibble: 15 × 4
## country import export product
## <chr> <dbl> <dbl> <chr>
## 1 Bolivia 46 0 N
## 2 Brazil 74 0 N
## 3 Chile 89 16 N
## 4 Colombia 77 16 A
## 5 CostaRica 84 21 A
## 6 Cuba 89 15 A
## 7 Ecuador 70 6 A
## 8 ElSalvador 60 13 A
## 9 Guatemala 55 9 A
## 10 Haiti 35 3 A
## 11 Jamaica 87 23 A
## 12 Mexico 83 4 N
## 13 Nicaragua 68 0 A
## 14 Paraguay 74 3 A
## 15 TrinidadTobago 84 15 AReplacing NAs
Because R will automatically fill any missing data with NAs,
sometimes we want to replace NA values with another value. For example
if blank cells actually = 0. We can use replace()with
is.na, or a special variation of the replace()
function, replace_na() to do this.
# Replace NAs ----------------------
# specifying columns
# first check which columns have NA values
is.na(missing.df) # import and export are the only columns with NAs## country import export product
## [1,] FALSE FALSE FALSE FALSE
## [2,] FALSE FALSE FALSE FALSE
## [3,] FALSE FALSE FALSE FALSE
## [4,] FALSE FALSE FALSE FALSE
## [5,] FALSE FALSE FALSE FALSE
## [6,] FALSE FALSE FALSE FALSE
## [7,] FALSE TRUE FALSE FALSE
## [8,] FALSE FALSE FALSE FALSE
## [9,] FALSE FALSE FALSE FALSE
## [10,] FALSE FALSE FALSE FALSE
## [11,] FALSE FALSE FALSE FALSE
## [12,] FALSE FALSE TRUE FALSE
## [13,] FALSE FALSE FALSE FALSE
## [14,] FALSE FALSE FALSE FALSE
## [15,] FALSE FALSE FALSE FALSE
## [16,] FALSE TRUE FALSE FALSE
## [17,] FALSE FALSE FALSE FALSE
## [18,] FALSE FALSE TRUE FALSE
## [19,] FALSE FALSE FALSE FALSE
## [20,] FALSE TRUE FALSE FALSE# replace NA in import with zeros using mutate() and replace_na()
missing.df %>%
# replace NA with 0
mutate(import = replace_na(import, 0),
export = replace_na(export, 0)) # provide the variable/column name and then what you want the NAs replaced with (e.g. 0)## # A tibble: 20 × 4
## country import export product
## <chr> <dbl> <dbl> <chr>
## 1 Bolivia 46 0 N
## 2 Brazil 74 0 N
## 3 Chile 89 16 N
## 4 Colombia 77 16 A
## 5 CostaRica 84 21 A
## 6 Cuba 89 15 A
## 7 DominicanRep 0 14 A
## 8 Ecuador 70 6 A
## 9 ElSalvador 60 13 A
## 10 Guatemala 55 9 A
## 11 Haiti 35 3 A
## 12 Honduras 51 0 A
## 13 Jamaica 87 23 A
## 14 Mexico 83 4 N
## 15 Nicaragua 68 0 A
## 16 Panama 0 19 N
## 17 Paraguay 74 3 A
## 18 Peru 73 0 N
## 19 TrinidadTobago 84 15 A
## 20 Venezuela 0 7 N# or using tidyverse trickery (less code repetition)
missing.df %>%
# replace NA w/ 0
mutate(across(
where(is.numeric),
~ replace_na(., 0)))## # A tibble: 20 × 4
## country import export product
## <chr> <dbl> <dbl> <chr>
## 1 Bolivia 46 0 N
## 2 Brazil 74 0 N
## 3 Chile 89 16 N
## 4 Colombia 77 16 A
## 5 CostaRica 84 21 A
## 6 Cuba 89 15 A
## 7 DominicanRep 0 14 A
## 8 Ecuador 70 6 A
## 9 ElSalvador 60 13 A
## 10 Guatemala 55 9 A
## 11 Haiti 35 3 A
## 12 Honduras 51 0 A
## 13 Jamaica 87 23 A
## 14 Mexico 83 4 N
## 15 Nicaragua 68 0 A
## 16 Panama 0 19 N
## 17 Paraguay 74 3 A
## 18 Peru 73 0 N
## 19 TrinidadTobago 84 15 A
## 20 Venezuela 0 7 N# for entire data set
# replace NA in missing.df with zeros using replace()
missing.df %>%
# replace NW w/ 0
replace(is.na(.), 0)## # A tibble: 20 × 4
## country import export product
## <chr> <dbl> <dbl> <chr>
## 1 Bolivia 46 0 N
## 2 Brazil 74 0 N
## 3 Chile 89 16 N
## 4 Colombia 77 16 A
## 5 CostaRica 84 21 A
## 6 Cuba 89 15 A
## 7 DominicanRep 0 14 A
## 8 Ecuador 70 6 A
## 9 ElSalvador 60 13 A
## 10 Guatemala 55 9 A
## 11 Haiti 35 3 A
## 12 Honduras 51 0 A
## 13 Jamaica 87 23 A
## 14 Mexico 83 4 N
## 15 Nicaragua 68 0 A
## 16 Panama 0 19 N
## 17 Paraguay 74 3 A
## 18 Peru 73 0 N
## 19 TrinidadTobago 84 15 A
## 20 Venezuela 0 7 NWe could also replace all values with a value calculated from the existing column data (e.g., mean, median, etc.)
# Replace all missing values in the data frame with the mean for the column
# check what the mean is for import and export columns
summary(missing.df)## country import export product
## Length:20 Min. :35.00 Min. : 0.00 Length:20
## Class :character 1st Qu.:60.00 1st Qu.: 3.25 Class :character
## Mode :character Median :74.00 Median :11.00 Mode :character
## Mean :70.53 Mean :10.22
## 3rd Qu.:84.00 3rd Qu.:15.75
## Max. :89.00 Max. :23.00
## NA's :3 NA's :2missing.df %>%
# replace NA with mean value for each column
mutate(export = replace_na(export,
mean(export,
na.rm = T)),
import = replace_na(import,
mean(import,
na.rm = T)))## # A tibble: 20 × 4
## country import export product
## <chr> <dbl> <dbl> <chr>
## 1 Bolivia 46 0 N
## 2 Brazil 74 0 N
## 3 Chile 89 16 N
## 4 Colombia 77 16 A
## 5 CostaRica 84 21 A
## 6 Cuba 89 15 A
## 7 DominicanRep 70.5 14 A
## 8 Ecuador 70 6 A
## 9 ElSalvador 60 13 A
## 10 Guatemala 55 9 A
## 11 Haiti 35 3 A
## 12 Honduras 51 10.2 A
## 13 Jamaica 87 23 A
## 14 Mexico 83 4 N
## 15 Nicaragua 68 0 A
## 16 Panama 70.5 19 N
## 17 Paraguay 74 3 A
## 18 Peru 73 10.2 N
## 19 TrinidadTobago 84 15 A
## 20 Venezuela 70.5 7 N# or using tidyverse trickery (less code repetition)
missing.df %>%
# replace NA with mean value for each column
mutate(across(
where(is.numeric),
~ replace_na(.,
mean(.,
na.rm=T))))## # A tibble: 20 × 4
## country import export product
## <chr> <dbl> <dbl> <chr>
## 1 Bolivia 46 0 N
## 2 Brazil 74 0 N
## 3 Chile 89 16 N
## 4 Colombia 77 16 A
## 5 CostaRica 84 21 A
## 6 Cuba 89 15 A
## 7 DominicanRep 70.5 14 A
## 8 Ecuador 70 6 A
## 9 ElSalvador 60 13 A
## 10 Guatemala 55 9 A
## 11 Haiti 35 3 A
## 12 Honduras 51 10.2 A
## 13 Jamaica 87 23 A
## 14 Mexico 83 4 N
## 15 Nicaragua 68 0 A
## 16 Panama 70.5 19 N
## 17 Paraguay 74 3 A
## 18 Peru 73 10.2 N
## 19 TrinidadTobago 84 15 A
## 20 Venezuela 70.5 7 NThere are tons of ways to replace NAs in data and we don’t have time to cover them all. Here is a link with more ways to replace NAs in data
Changing values to NA
What about the reverse? When we have values in our data that are
meant to be NAs. This happens frequently, we type ‘none’, ‘n/a’, ‘-’,
etc. instead of NA and R reads those in as characters
instead of missing data. We’ve already learned a few ways to do this
with mutate(). Let’s review with a new data set.
Download the bobcat necropsy data (Bobcat_necropsy_data.csv) and save it to the data folder in your project directory.
In the same code chunk complete the following steps
Read in the data and save it as bobcats
Set the column names to lowercase
Select only ‘necropsy’, ‘necropsydata’, ‘age’, and ‘sex’
print a summary of the data
# Replace values with NA ----------------------
# FILL IN YOUR CODE HERE ----------------------
bobcats <- read_csv('data/raw/Bobcat_necropsy_data.csv') %>%
# set names to lowercase
set_names(
names(.) %>%
tolower()) %>%
# select specific columns
select(necropsy, necropsydate, age, sex)
summary(bobcats)## necropsy necropsydate age sex
## Min. : 1 Length:121 Length:121 Length:121
## 1st Qu.: 31 Class :character Class :character Class :character
## Median : 61 Mode :character Mode :character Mode :character
## Mean : 61
## 3rd Qu.: 91
## Max. :121Now using the mutate()
function set age as a numeric
variable.
bobcats %>%
# set age to numeric
mutate(age = as.numeric(age))## # A tibble: 121 × 4
## necropsy necropsydate age sex
## <dbl> <chr> <dbl> <chr>
## 1 1 3/6/19 3 M
## 2 2 3/9/19 1 F
## 3 3 3/10/19 2 M
## 4 4 3/13/19 0 F
## 5 5 3/23/19 1 M
## 6 6 3/24/19 NA M
## 7 7 3/31/19 NA M
## 8 8 4/7/19 1 M
## 9 9 4/13/19 0 M
## 10 10 4/14/19 3 M
## # ℹ 111 more rowsWhen we changed ‘aged’ to a numeric variable this
automatically change non-numeric entries to NAs, but if we were working
with a character instead we could change some entries
to NA using mutate() and replace(). Replace ‘na’ entries in the ‘age’ column
usingmutate() and replace() or
recode().
# replace 'na' in age using replace function
bobcats %>%
mutate(age = replace(age,
age == 'na',
NA))## # A tibble: 121 × 4
## necropsy necropsydate age sex
## <dbl> <chr> <chr> <chr>
## 1 1 3/6/19 3 M
## 2 2 3/9/19 1 F
## 3 3 3/10/19 2 M
## 4 4 3/13/19 0 F
## 5 5 3/23/19 1 M
## 6 6 3/24/19 X M
## 7 7 3/31/19 X M
## 8 8 4/7/19 1 M
## 9 9 4/13/19 0 M
## 10 10 4/14/19 3 M
## # ℹ 111 more rows# replace na in age using recode
bobcats %>%
mutate(age = recode(age,
'na' = NA))## # A tibble: 121 × 4
## necropsy necropsydate age sex
## <dbl> <chr> <lgl> <chr>
## 1 1 3/6/19 NA M
## 2 2 3/9/19 NA F
## 3 3 3/10/19 NA M
## 4 4 3/13/19 NA F
## 5 5 3/23/19 NA M
## 6 6 3/24/19 NA M
## 7 7 3/31/19 NA M
## 8 8 4/7/19 NA M
## 9 9 4/13/19 NA M
## 10 10 4/14/19 NA M
## # ℹ 111 more rowsBut you may notice there are still some questionable entries in the age column. X has been entered in some places and this should also be an NA. We can handle changing multiple entries by creating a vectors with the strings (characters) we want to replace
# change multiple entries to NA
# make a vector with a string of values that are meant to be NAs in data
na_string <- c('na', 'X')
# use mutate with %in% argument to replace all values in the string above with NA
bobcats %>%
mutate(age = replace(age,
age %in% na_string,
NA))## # A tibble: 121 × 4
## necropsy necropsydate age sex
## <dbl> <chr> <chr> <chr>
## 1 1 3/6/19 3 M
## 2 2 3/9/19 1 F
## 3 3 3/10/19 2 M
## 4 4 3/13/19 0 F
## 5 5 3/23/19 1 M
## 6 6 3/24/19 <NA> M
## 7 7 3/31/19 <NA> M
## 8 8 4/7/19 1 M
## 9 9 4/13/19 0 M
## 10 10 4/14/19 3 M
## # ℹ 111 more rowsPivot data
Recall that the tidyverse packages are all designed to work with tidy data. The tidyr package has several functions to help you get your data into this format.
Use “pivot_longer()” to get all of the values and variables from multiple columns into a single column.
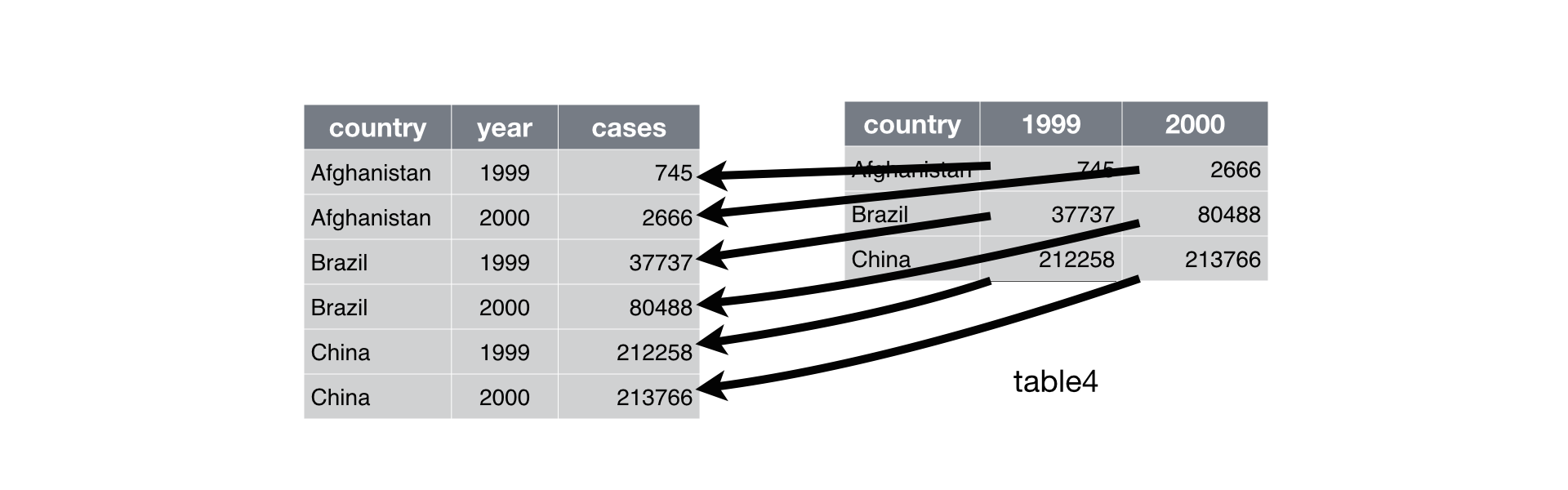 Use “pivot_wider()” to
distribute two variables in a single column into separate columns, with
their data values(‘value’)
Use “pivot_wider()” to
distribute two variables in a single column into separate columns, with
their data values(‘value’)
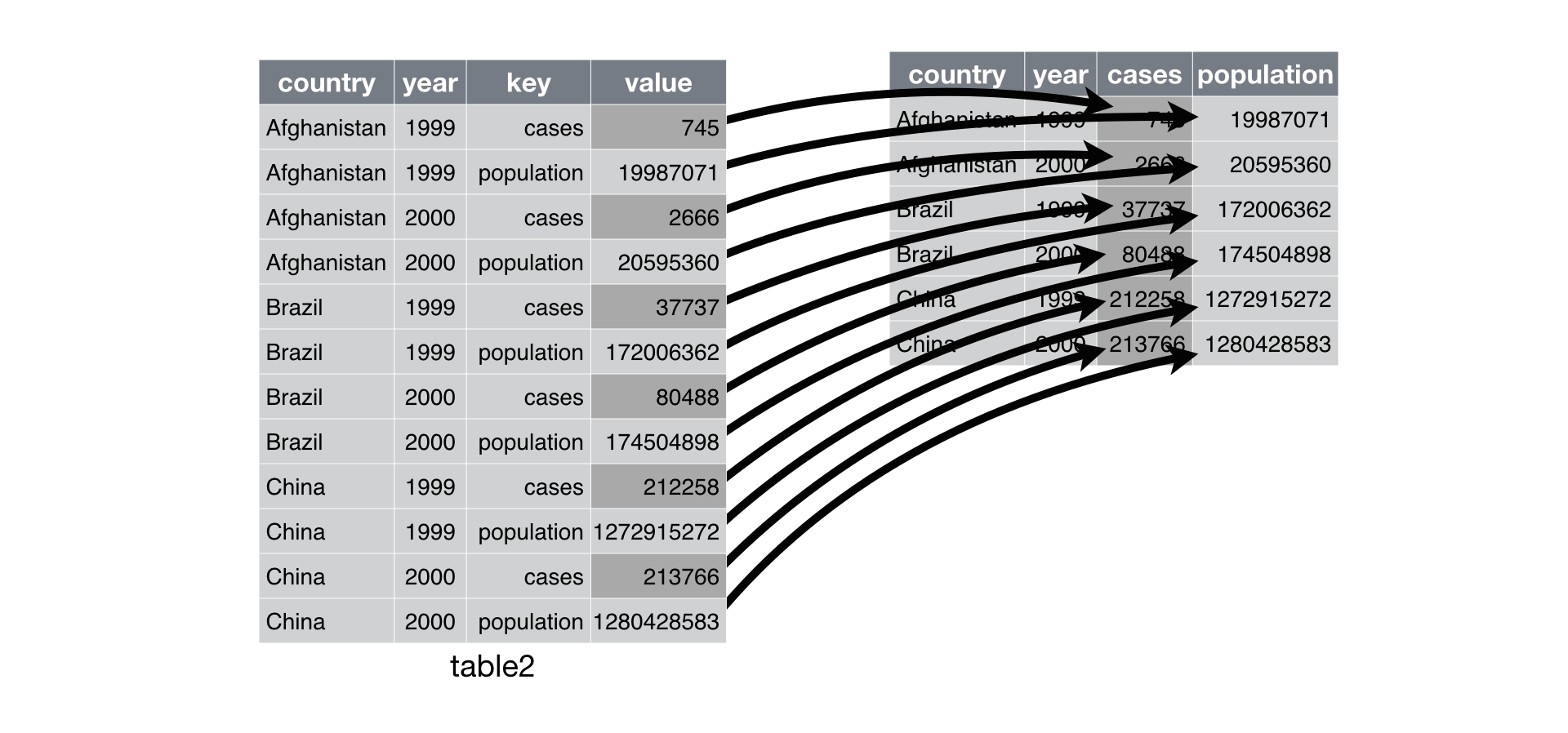 Let’s go through an example
using
Let’s go through an example
using pivot_longer() and data from the tidyr
package.
# Pivot functions ----------------------
# use data from tidyr package
relig_income## # A tibble: 18 × 11
## religion `<$10k` `$10-20k` `$20-30k` `$30-40k` `$40-50k` `$50-75k` `$75-100k`
## <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 Agnostic 27 34 60 81 76 137 122
## 2 Atheist 12 27 37 52 35 70 73
## 3 Buddhist 27 21 30 34 33 58 62
## 4 Catholic 418 617 732 670 638 1116 949
## 5 Don’t k… 15 14 15 11 10 35 21
## 6 Evangel… 575 869 1064 982 881 1486 949
## 7 Hindu 1 9 7 9 11 34 47
## 8 Histori… 228 244 236 238 197 223 131
## 9 Jehovah… 20 27 24 24 21 30 15
## 10 Jewish 19 19 25 25 30 95 69
## 11 Mainlin… 289 495 619 655 651 1107 939
## 12 Mormon 29 40 48 51 56 112 85
## 13 Muslim 6 7 9 10 9 23 16
## 14 Orthodox 13 17 23 32 32 47 38
## 15 Other C… 9 7 11 13 13 14 18
## 16 Other F… 20 33 40 46 49 63 46
## 17 Other W… 5 2 3 4 2 7 3
## 18 Unaffil… 217 299 374 365 341 528 407
## # ℹ 3 more variables: `$100-150k` <dbl>, `>150k` <dbl>,
## # `Don't know/refused` <dbl>relig_income %>%
pivot_longer(!religion, names_to = "income", values_to = "count")## # A tibble: 180 × 3
## religion income count
## <chr> <chr> <dbl>
## 1 Agnostic <$10k 27
## 2 Agnostic $10-20k 34
## 3 Agnostic $20-30k 60
## 4 Agnostic $30-40k 81
## 5 Agnostic $40-50k 76
## 6 Agnostic $50-75k 137
## 7 Agnostic $75-100k 122
## 8 Agnostic $100-150k 109
## 9 Agnostic >150k 84
## 10 Agnostic Don't know/refused 96
## # ℹ 170 more rowsJoining data
Sometimes data you need for a singular figure or analysis have been entered in two separate data files. We can join data from matching observations (e.g., same individual, site, etc.) using join functions in dplyr based on the keys (columns) in the data.
Here’s a visual representation of how the various join functions work.

Create fake data
First, let’s get some practice with a few functions we learned at the beginning of the day to create some fake environmental data for the turtles data set.
# Joining data----------------------
# create some fake data first
turtles_env <- tibble(
# create variable 'tag' ranging from 1 - 105
tag = 1:105,
# create variable site with 3 sites that repeat for 35 indv each
site = rep(c('A', 'B', 'C'),
each = 35),
# create a variable for average summer temperature in Celsius
avg_summer_temp = runif(105,
min = 15,
max = 30),
# create a variable for average summer precipitation in mm
avg_summer_precip = runif(105,
min = 0,
max = 110)
)
turtles_env## # A tibble: 105 × 4
## tag site avg_summer_temp avg_summer_precip
## <int> <chr> <dbl> <dbl>
## 1 1 A 24.8 13.4
## 2 2 A 26.0 48.4
## 3 3 A 21.3 80.5
## 4 4 A 23.2 101.
## 5 5 A 19.4 87.1
## 6 6 A 20.4 51.5
## 7 7 A 26.2 23.6
## 8 8 A 16.4 22.7
## 9 9 A 23.2 88.1
## 10 10 A 25.9 7.00
## # ℹ 95 more rowsNow we can join the environmental data we just created with the turtle observation data (turtles_tidy) using the tag column.
Left join
Let’s start by using left_join() to join the two
datasets together.
# Left join----------------------
turtles_full <- turtles_tidy %>%
# left join with turtles_env
left_join(turtles_env,
by = 'tag')
turtles_full## # A tibble: 21 × 8
## tag sex c_length h_width weight site avg_summer_temp avg_summer_precip
## <dbl> <fct> <dbl> <dbl> <dbl> <chr> <dbl> <dbl>
## 1 10 male 41 7.15 7.6 A 25.9 7.00
## 2 11 female 46.4 8.18 11 A 15.1 12.4
## 3 2 <NA> 24.3 4.42 1.65 A 26.0 48.4
## 4 15 <NA> 28.7 4.89 2.18 A 18.4 76.3
## 5 16 <NA> 32 5.37 3 A 18.9 16.2
## 6 3 female 42.8 7.32 8.6 A 21.3 80.5
## 7 4 male 40 6.6 6.5 A 23.2 101.
## 8 5 female 45 8.05 10.9 A 19.4 87.1
## 9 12 female 44 7.55 8.9 A 23.0 11.0
## 10 13 <NA> 28 4.85 1.97 A 28.4 28.9
## # ℹ 11 more rowsNotice how are dataset reamins the same length as the turtles_tidy
dataset and all environmental observations that for tags not in
the turtles_tidy dataset are dropped. This is how
left_join() works.
If we wanted to keep all the environmental data and add the observation data we could use right join.
Right join
# Right join ----------------------
turtles_tidy %>%
# right join
right_join(turtles_env,
by = 'tag') %>%
# reorder by tag number so we can see what happened
arrange(desc(tag))## # A tibble: 105 × 8
## tag sex c_length h_width weight site avg_summer_temp avg_summer_precip
## <dbl> <fct> <dbl> <dbl> <dbl> <chr> <dbl> <dbl>
## 1 105 male 44 7.1 9 C 17.0 22.2
## 2 104 male 44 7.35 9 C 17.1 31.9
## 3 103 <NA> NA NA NA C 19.5 19.3
## 4 102 <NA> NA NA NA C 20.8 60.6
## 5 101 <NA> NA NA NA C 29.5 12.5
## 6 100 <NA> NA NA NA C 26.4 96.5
## 7 99 <NA> NA NA NA C 16.3 42.3
## 8 98 <NA> NA NA NA C 15.2 55.8
## 9 97 <NA> NA NA NA C 28.0 9.87
## 10 96 <NA> NA NA NA C 29.6 23.8
## # ℹ 95 more rowsNow, wherever we don’t have turtle observation data to match the environmental data we get NAs
Inner join
The inner_join() function will join values from both
datasets only where there are observations for both. With our current
dataset this would perform the same as left_join() because
there aren’t any missing data for the turtles_env dataset. But if we
adjust that dataset we can see the difference.
# Inner join ----------------------
# run inner join without altering data to see what happen
turtles_tidy %>%
inner_join(turtles_env,
by = 'tag')## # A tibble: 21 × 8
## tag sex c_length h_width weight site avg_summer_temp avg_summer_precip
## <dbl> <fct> <dbl> <dbl> <dbl> <chr> <dbl> <dbl>
## 1 10 male 41 7.15 7.6 A 25.9 7.00
## 2 11 female 46.4 8.18 11 A 15.1 12.4
## 3 2 <NA> 24.3 4.42 1.65 A 26.0 48.4
## 4 15 <NA> 28.7 4.89 2.18 A 18.4 76.3
## 5 16 <NA> 32 5.37 3 A 18.9 16.2
## 6 3 female 42.8 7.32 8.6 A 21.3 80.5
## 7 4 male 40 6.6 6.5 A 23.2 101.
## 8 5 female 45 8.05 10.9 A 19.4 87.1
## 9 12 female 44 7.55 8.9 A 23.0 11.0
## 10 13 <NA> 28 4.85 1.97 A 28.4 28.9
## # ℹ 11 more rows# if we remove some observations (say we didn't collect environmental data for the first 10 captures) then we can see the diff
turtles_env.sub <- turtles_env %>%
# filter to only tags greater than 10
filter(tag > 10)
head(turtles_env.sub)## # A tibble: 6 × 4
## tag site avg_summer_temp avg_summer_precip
## <int> <chr> <dbl> <dbl>
## 1 11 A 15.1 12.4
## 2 12 A 23.0 11.0
## 3 13 A 28.4 28.9
## 4 14 A 24.0 27.7
## 5 15 A 18.4 76.3
## 6 16 A 18.9 16.2# now join with turtles_tidy
turtles_tidy %>%
inner_join(turtles_env.sub,
by = 'tag')## # A tibble: 11 × 8
## tag sex c_length h_width weight site avg_summer_temp avg_summer_precip
## <dbl> <fct> <dbl> <dbl> <dbl> <chr> <dbl> <dbl>
## 1 11 female 46.4 8.18 11 A 15.1 12.4
## 2 15 <NA> 28.7 4.89 2.18 A 18.4 76.3
## 3 16 <NA> 32 5.37 3 A 18.9 16.2
## 4 12 female 44 7.55 8.9 A 23.0 11.0
## 5 13 <NA> 28 4.85 1.97 A 28.4 28.9
## 6 17 female 35.1 6.04 4.5 A 24.7 11.5
## 7 19 male 42.3 6.77 7.8 A 18.1 109.
## 8 22 female 48.1 8.55 12.8 A 15.8 1.80
## 9 105 male 44 7.1 9 C 17.0 22.2
## 10 14 male 43 6.6 7.2 A 24.0 27.7
## 11 104 male 44 7.35 9 C 17.1 31.9# notice fewer rowsFull join
Lastly we will look at full_join(), which does exactly
as the name suggests, it joins all the data even if there aren’t matches
between the datasets
# Full join ----------------------
turtles_tidy %>%
full_join(turtles_env,
by = 'tag')## # A tibble: 105 × 8
## tag sex c_length h_width weight site avg_summer_temp avg_summer_precip
## <dbl> <fct> <dbl> <dbl> <dbl> <chr> <dbl> <dbl>
## 1 10 male 41 7.15 7.6 A 25.9 7.00
## 2 11 female 46.4 8.18 11 A 15.1 12.4
## 3 2 <NA> 24.3 4.42 1.65 A 26.0 48.4
## 4 15 <NA> 28.7 4.89 2.18 A 18.4 76.3
## 5 16 <NA> 32 5.37 3 A 18.9 16.2
## 6 3 female 42.8 7.32 8.6 A 21.3 80.5
## 7 4 male 40 6.6 6.5 A 23.2 101.
## 8 5 female 45 8.05 10.9 A 19.4 87.1
## 9 12 female 44 7.55 8.9 A 23.0 11.0
## 10 13 <NA> 28 4.85 1.97 A 28.4 28.9
## # ℹ 95 more rowsBut what if you have different variable names for your key (e.g., tag). Well you could change the variable name in one of the datasets so they match, but you can also specify what the keys in each dataset are called
Mismatched keys
# Mismatched keys----------------------
# rename tag in the turtles_env dataset
turtles_env <- turtles_env %>%
rename(tag_number = tag)
names(turtles_env)## [1] "tag_number" "site" "avg_summer_temp"
## [4] "avg_summer_precip"# now join with mismatched names
turtles_tidy %>%
left_join(turtles_env,
join_by('tag' == 'tag_number'))## # A tibble: 21 × 8
## tag sex c_length h_width weight site avg_summer_temp avg_summer_precip
## <dbl> <fct> <dbl> <dbl> <dbl> <chr> <dbl> <dbl>
## 1 10 male 41 7.15 7.6 A 25.9 7.00
## 2 11 female 46.4 8.18 11 A 15.1 12.4
## 3 2 <NA> 24.3 4.42 1.65 A 26.0 48.4
## 4 15 <NA> 28.7 4.89 2.18 A 18.4 76.3
## 5 16 <NA> 32 5.37 3 A 18.9 16.2
## 6 3 female 42.8 7.32 8.6 A 21.3 80.5
## 7 4 male 40 6.6 6.5 A 23.2 101.
## 8 5 female 45 8.05 10.9 A 19.4 87.1
## 9 12 female 44 7.55 8.9 A 23.0 11.0
## 10 13 <NA> 28 4.85 1.97 A 28.4 28.9
## # ℹ 11 more rowsPractice Problems
1 Remove NAs
- Read in the turtles_tidy data
- In the same code chunk remove all rows with NAs
- Assign this new data to the environment as “turtles_no_na”
## # A tibble: 15 × 5
## tag sex c_length h_width weight
## <dbl> <fct> <dbl> <dbl> <dbl>
## 1 10 male 41 7.15 7.6
## 2 11 female 46.4 8.18 11
## 3 3 female 42.8 7.32 8.6
## 4 4 male 40 6.6 6.5
## 5 5 female 45 8.05 10.9
## 6 12 female 44 7.55 8.9
## 7 6 female 40 6.53 6.2
## 8 9 male 35 5.74 3.9
## 9 17 female 35.1 6.04 4.5
## 10 19 male 42.3 6.77 7.8
## 11 22 female 48.1 8.55 12.8
## 12 105 male 44 7.1 9
## 13 14 male 43 6.6 7.2
## 14 7 female 48 8.67 13.5
## 15 104 male 44 7.35 92 Make a new variable
Using the turtles_no_na data, overwrite the data and make a new
variable called “size_class” based on the “weight” variable using
case_when() whereby
weights less than 4 are juvenile
weights greater than 7 are adult
weights between 4 and 7 are subadult
(There are multiple ways to do this which is why there are multiple printouts, but they will yield the same answer)
## [1] "adult" "adult" "adult" "subadult" "adult" "adult"
## [7] "subadult" "juvenile" "subadult" "adult" "adult" "adult"
## [13] "adult" "adult" "adult"## [1] "adult" "adult" "adult" "subadult" "adult" "adult"
## [7] "subadult" "juvenile" "subadult" "adult" "adult" "adult"
## [13] "adult" "adult" "adult"3 Replace values with NA
In the turtles_tidy data (not the turtles_no_na data) replace ALL variable values for tags 104 and 105 with NAs
Hint you will need to create a vector for the tag numbers you
want to replace and use mutate()
## # A tibble: 21 × 5
## tag sex c_length h_width weight
## <dbl> <fct> <dbl> <dbl> <dbl>
## 1 10 male 41 7.15 7.6
## 2 11 female 46.4 8.18 11
## 3 2 <NA> 24.3 4.42 1.65
## 4 15 <NA> 28.7 4.89 2.18
## 5 16 <NA> 32 5.37 3
## 6 3 female 42.8 7.32 8.6
## 7 4 male 40 6.6 6.5
## 8 5 female 45 8.05 10.9
## 9 12 female 44 7.55 8.9
## 10 13 <NA> 28 4.85 1.97
## # ℹ 11 more rows## # A tibble: 21 × 5
## tag sex c_length h_width weight
## <dbl> <fct> <dbl> <dbl> <dbl>
## 1 10 male 41 7.15 7.6
## 2 11 female 46.4 8.18 11
## 3 2 <NA> 24.3 4.42 1.65
## 4 15 <NA> 28.7 4.89 2.18
## 5 16 <NA> 32 5.37 3
## 6 3 female 42.8 7.32 8.6
## 7 4 male 40 6.6 6.5
## 8 5 female 45 8.05 10.9
## 9 12 female 44 7.55 8.9
## 10 13 <NA> 28 4.85 1.97
## # ℹ 11 more rows4 Pivot data
Use the below code to read in the Soils data from the carData package
# Load the example data
soil <- carData::Soils # load example dataprint the first few lines of data in “soil”
Pivot the data so that columns Ca - Na are contained in one column called nutrients (again there are two possible solutions (really more than that but two I expect people to use))
## Group Contour Depth Gp Block pH N Dens P Ca Mg K Na Conduc
## 1 1 Top 0-10 T0 1 5.40 0.188 0.92 215 16.35 7.65 0.72 1.14 1.09
## 2 1 Top 0-10 T0 2 5.65 0.165 1.04 208 12.25 5.15 0.71 0.94 1.35
## 3 1 Top 0-10 T0 3 5.14 0.260 0.95 300 13.02 5.68 0.68 0.60 1.41
## 4 1 Top 0-10 T0 4 5.14 0.169 1.10 248 11.92 7.88 1.09 1.01 1.64
## 5 2 Top 10-30 T1 1 5.14 0.164 1.12 174 14.17 8.12 0.70 2.17 1.85
## 6 2 Top 10-30 T1 2 5.10 0.094 1.22 129 8.55 6.92 0.81 2.67 3.18## # A tibble: 192 × 12
## Group Contour Depth Gp Block pH N Dens P Conduc nutrient value
## <fct> <fct> <fct> <fct> <fct> <dbl> <dbl> <dbl> <int> <dbl> <chr> <dbl>
## 1 1 Top 0-10 T0 1 5.4 0.188 0.92 215 1.09 Ca 16.4
## 2 1 Top 0-10 T0 1 5.4 0.188 0.92 215 1.09 Mg 7.65
## 3 1 Top 0-10 T0 1 5.4 0.188 0.92 215 1.09 K 0.72
## 4 1 Top 0-10 T0 1 5.4 0.188 0.92 215 1.09 Na 1.14
## 5 1 Top 0-10 T0 2 5.65 0.165 1.04 208 1.35 Ca 12.2
## 6 1 Top 0-10 T0 2 5.65 0.165 1.04 208 1.35 Mg 5.15
## 7 1 Top 0-10 T0 2 5.65 0.165 1.04 208 1.35 K 0.71
## 8 1 Top 0-10 T0 2 5.65 0.165 1.04 208 1.35 Na 0.94
## 9 1 Top 0-10 T0 3 5.14 0.26 0.95 300 1.41 Ca 13.0
## 10 1 Top 0-10 T0 3 5.14 0.26 0.95 300 1.41 Mg 5.68
## # ℹ 182 more rows## # A tibble: 192 × 12
## Group Contour Depth Gp Block pH N Dens P Conduc nutrient value
## <fct> <fct> <fct> <fct> <fct> <dbl> <dbl> <dbl> <int> <dbl> <chr> <dbl>
## 1 1 Top 0-10 T0 1 5.4 0.188 0.92 215 1.09 Ca 16.4
## 2 1 Top 0-10 T0 1 5.4 0.188 0.92 215 1.09 Mg 7.65
## 3 1 Top 0-10 T0 1 5.4 0.188 0.92 215 1.09 K 0.72
## 4 1 Top 0-10 T0 1 5.4 0.188 0.92 215 1.09 Na 1.14
## 5 1 Top 0-10 T0 2 5.65 0.165 1.04 208 1.35 Ca 12.2
## 6 1 Top 0-10 T0 2 5.65 0.165 1.04 208 1.35 Mg 5.15
## 7 1 Top 0-10 T0 2 5.65 0.165 1.04 208 1.35 K 0.71
## 8 1 Top 0-10 T0 2 5.65 0.165 1.04 208 1.35 Na 0.94
## 9 1 Top 0-10 T0 3 5.14 0.26 0.95 300 1.41 Ca 13.0
## 10 1 Top 0-10 T0 3 5.14 0.26 0.95 300 1.41 Mg 5.68
## # ℹ 182 more rows5 Join Data
If you haven’t already download the 3 bobcat data files added to the course after 12 January 2024 and save them to the data/raw folder
Bobcat collection data for Purrr (bobcat_collection_data.csv)
Bobcat necropsy data for Purrr (bobcat_necropsy_only_data.csv)
Bobcat age data for Purrr (bobcat_age_data.csv)
- Read in the data files using the tidyverse function
- In the same code chunk, set the column names to lowercase for all 3
data sets AND rename the ‘Bobcat_ID#’ column to bobcat_id (NOTE:
this requires a lot of code repition which is annoying and does not
follow best coding practices, we will learn a much better way to do this
when we cover Purrr)
- Use the csv file names as the object names when you assign them to
the environment - Make a list with the three data sets and check their
internal structure (there are multiple ways to do this)
- Join the bobcat_necropsy_only_data to the
bobcat_collection_data AND then in the same code chunk join the
bobcat_age_data as well. Make sure to retain all observations from the
bobcat_collection_data. You will need to use the bobcat_id column as
the key when joining
- Print the summary of your data to check that it worked
## List of 3
## $ : spc_tbl_ [121 × 7] (S3: spec_tbl_df/tbl_df/tbl/data.frame)
## ..$ bobcat_id : chr [1:121] "5/18/01" "5/18/02" "58-18-03" "64-19-04" ...
## ..$ county : chr [1:121] "Athens" "Athens" "Morgan" "Perry" ...
## ..$ township : chr [1:121] "Dover" "Canaan" "Hower" "Reading" ...
## ..$ collectiondate: chr [1:121] "12/30/18" "12/14/18" "12/7/18" "2/10/19" ...
## ..$ month : num [1:121] 12 12 12 2 1 3 2 2 2 3 ...
## ..$ coordinates_n : num [1:121] 39.4 39.3 39.5 39.8 40.4 ...
## ..$ coordinates_w : num [1:121] -82.1 -82 -82 -82.3 -81.2 ...
## ..- attr(*, "spec")=
## .. .. cols(
## .. .. `Bobcat_ID#` = col_character(),
## .. .. County = col_character(),
## .. .. Township = col_character(),
## .. .. CollectionDate = col_character(),
## .. .. Month = col_double(),
## .. .. Coordinates_N = col_double(),
## .. .. Coordinates_W = col_double()
## .. .. )
## ..- attr(*, "problems")=<externalptr>
## $ : spc_tbl_ [121 × 14] (S3: spec_tbl_df/tbl_df/tbl/data.frame)
## ..$ bobcat_id : chr [1:121] "5/18/01" "5/18/02" "58-18-03" "64-19-04" ...
## ..$ necropsy : num [1:121] 1 2 3 4 5 6 7 8 9 10 ...
## ..$ necropsydate : chr [1:121] "3/6/19" "3/9/19" "3/10/19" "3/13/19" ...
## ..$ dissector : chr [1:121] "SP,_MD" "SP_MD_CH" "MD_CH" "MD_CH_HK" ...
## ..$ approxage : chr [1:121] "Ad" "Ad" "Ad" "Juv" ...
## ..$ sex : chr [1:121] "M" "F" "M" "F" ...
## ..$ fecundity_females: chr [1:121] "na" "na" "na" "0" ...
## ..$ rearfoot_cm : chr [1:121] "17.9" "16" "17.1" "14.7" ...
## ..$ tail_cm : chr [1:121] "11.5" "11.5" "11.4" "11.3" ...
## ..$ ear_cm : chr [1:121] "6.5" "6.5" "6.4" "6.5" ...
## ..$ body_w/tail_cm : chr [1:121] "89.5" "82" "92" "71.5" ...
## ..$ body : chr [1:121] "78" "70.5" "80.6" "60.2" ...
## ..$ weight_kg : chr [1:121] "13.6" "6.33" "9.98" "4.62" ...
## ..$ condition : chr [1:121] "17.43589744" "8.978723404" "12.382134" "7.674418605" ...
## ..- attr(*, "spec")=
## .. .. cols(
## .. .. `Bobcat_ID#` = col_character(),
## .. .. Necropsy = col_double(),
## .. .. NecropsyDate = col_character(),
## .. .. Dissector = col_character(),
## .. .. ApproxAge = col_character(),
## .. .. Sex = col_character(),
## .. .. Fecundity_Females = col_character(),
## .. .. RearFoot_cm = col_character(),
## .. .. Tail_cm = col_character(),
## .. .. Ear_cm = col_character(),
## .. .. `Body_w/Tail_cm` = col_character(),
## .. .. Body = col_character(),
## .. .. Weight_kg = col_character(),
## .. .. Condition = col_character()
## .. .. )
## ..- attr(*, "problems")=<externalptr>
## $ : spc_tbl_ [121 × 2] (S3: spec_tbl_df/tbl_df/tbl/data.frame)
## ..$ bobcat_id: chr [1:121] "5/18/01" "5/18/02" "58-18-03" "64-19-04" ...
## ..$ age : chr [1:121] "3" "1" "2" "0" ...
## ..- attr(*, "spec")=
## .. .. cols(
## .. .. `Bobcat_ID#` = col_character(),
## .. .. Age = col_character()
## .. .. )
## ..- attr(*, "problems")=<externalptr>## List of 3
## $ : spc_tbl_ [121 × 7] (S3: spec_tbl_df/tbl_df/tbl/data.frame)
## ..$ bobcat_id : chr [1:121] "5/18/01" "5/18/02" "58-18-03" "64-19-04" ...
## ..$ county : chr [1:121] "Athens" "Athens" "Morgan" "Perry" ...
## ..$ township : chr [1:121] "Dover" "Canaan" "Hower" "Reading" ...
## ..$ collectiondate: chr [1:121] "12/30/18" "12/14/18" "12/7/18" "2/10/19" ...
## ..$ month : num [1:121] 12 12 12 2 1 3 2 2 2 3 ...
## ..$ coordinates_n : num [1:121] 39.4 39.3 39.5 39.8 40.4 ...
## ..$ coordinates_w : num [1:121] -82.1 -82 -82 -82.3 -81.2 ...
## ..- attr(*, "spec")=
## .. .. cols(
## .. .. `Bobcat_ID#` = col_character(),
## .. .. County = col_character(),
## .. .. Township = col_character(),
## .. .. CollectionDate = col_character(),
## .. .. Month = col_double(),
## .. .. Coordinates_N = col_double(),
## .. .. Coordinates_W = col_double()
## .. .. )
## ..- attr(*, "problems")=<externalptr>
## $ : spc_tbl_ [121 × 14] (S3: spec_tbl_df/tbl_df/tbl/data.frame)
## ..$ bobcat_id : chr [1:121] "5/18/01" "5/18/02" "58-18-03" "64-19-04" ...
## ..$ necropsy : num [1:121] 1 2 3 4 5 6 7 8 9 10 ...
## ..$ necropsydate : chr [1:121] "3/6/19" "3/9/19" "3/10/19" "3/13/19" ...
## ..$ dissector : chr [1:121] "SP,_MD" "SP_MD_CH" "MD_CH" "MD_CH_HK" ...
## ..$ approxage : chr [1:121] "Ad" "Ad" "Ad" "Juv" ...
## ..$ sex : chr [1:121] "M" "F" "M" "F" ...
## ..$ fecundity_females: chr [1:121] "na" "na" "na" "0" ...
## ..$ rearfoot_cm : chr [1:121] "17.9" "16" "17.1" "14.7" ...
## ..$ tail_cm : chr [1:121] "11.5" "11.5" "11.4" "11.3" ...
## ..$ ear_cm : chr [1:121] "6.5" "6.5" "6.4" "6.5" ...
## ..$ body_w/tail_cm : chr [1:121] "89.5" "82" "92" "71.5" ...
## ..$ body : chr [1:121] "78" "70.5" "80.6" "60.2" ...
## ..$ weight_kg : chr [1:121] "13.6" "6.33" "9.98" "4.62" ...
## ..$ condition : chr [1:121] "17.43589744" "8.978723404" "12.382134" "7.674418605" ...
## ..- attr(*, "spec")=
## .. .. cols(
## .. .. `Bobcat_ID#` = col_character(),
## .. .. Necropsy = col_double(),
## .. .. NecropsyDate = col_character(),
## .. .. Dissector = col_character(),
## .. .. ApproxAge = col_character(),
## .. .. Sex = col_character(),
## .. .. Fecundity_Females = col_character(),
## .. .. RearFoot_cm = col_character(),
## .. .. Tail_cm = col_character(),
## .. .. Ear_cm = col_character(),
## .. .. `Body_w/Tail_cm` = col_character(),
## .. .. Body = col_character(),
## .. .. Weight_kg = col_character(),
## .. .. Condition = col_character()
## .. .. )
## ..- attr(*, "problems")=<externalptr>
## $ : spc_tbl_ [121 × 2] (S3: spec_tbl_df/tbl_df/tbl/data.frame)
## ..$ bobcat_id: chr [1:121] "5/18/01" "5/18/02" "58-18-03" "64-19-04" ...
## ..$ age : chr [1:121] "3" "1" "2" "0" ...
## ..- attr(*, "spec")=
## .. .. cols(
## .. .. `Bobcat_ID#` = col_character(),
## .. .. Age = col_character()
## .. .. )
## ..- attr(*, "problems")=<externalptr>## bobcat_id county township collectiondate
## Length:121 Length:121 Length:121 Length:121
## Class :character Class :character Class :character Class :character
## Mode :character Mode :character Mode :character Mode :character
##
##
##
## month coordinates_n coordinates_w necropsy
## Min. : 1.000 Min. :38.25 Min. :-83.25 Min. : 1
## 1st Qu.: 2.000 1st Qu.:39.25 1st Qu.:-82.38 1st Qu.: 31
## Median : 3.000 Median :39.59 Median :-81.95 Median : 61
## Mean : 5.826 Mean :39.58 Mean :-81.95 Mean : 61
## 3rd Qu.:11.000 3rd Qu.:39.97 3rd Qu.:-81.50 3rd Qu.: 91
## Max. :12.000 Max. :40.68 Max. :-80.89 Max. :121
## necropsydate dissector approxage sex
## Length:121 Length:121 Length:121 Length:121
## Class :character Class :character Class :character Class :character
## Mode :character Mode :character Mode :character Mode :character
##
##
##
## fecundity_females rearfoot_cm tail_cm ear_cm
## Length:121 Length:121 Length:121 Length:121
## Class :character Class :character Class :character Class :character
## Mode :character Mode :character Mode :character Mode :character
##
##
##
## body_w/tail_cm body weight_kg condition
## Length:121 Length:121 Length:121 Length:121
## Class :character Class :character Class :character Class :character
## Mode :character Mode :character Mode :character Mode :character
##
##
##
## age
## Length:121
## Class :character
## Mode :character
##
##
##